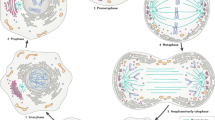
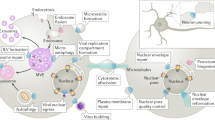
Abstract
Membrane trafficking and mitosis are two essential processes in eukaryotic cells. Surprisingly, many proteins best known for their role in membrane trafficking have additional 'moonlighting' functions in mitosis. Despite having proteins in common, there is insufficient evidence for a specific connection between these two processes. Instead, these phenomena demonstrate the adaptability of the membrane trafficking machinery that allows its repurposing for different cellular functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Beadle, G. W. & Tatum, E. L. Genetic control of biochemical reactions in Neurospora. Proc. Natl Acad. Sci. USA 27, 499–506 (1941).
Pan, Q., Shai, O., Lee, L. J., Frey, B. J. & Blencowe, B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genet. 40, 1413–1415 (2008).
Jeffery, C. J. Moonlighting proteins. Trends Biochem. Sci. 24, 8–11 (1999).
Jeffery, C. J. Moonlighting proteins: old proteins learning new tricks. Trends Genet. 19, 415–417 (2003).
Copley, S. D. Moonlighting is mainstream: paradigm adjustment required. Bioessays 34, 578–588 (2012).
Royle, S. J. The role of clathrin in mitotic spindle organisation. J. Cell Sci. 125, 19–28 (2012).
Maro, B., Johnson, M. H., Pickering, S. J. & Louvard, D. Changes in the distribution of membranous organelles during mouse early development. J. Embryol. Exp. Morphol. 90, 287–309 (1985).
Okamoto, C. T., McKinney, J. & Jeng, Y. Y. Clathrin in mitotic spindles. Am. J. Physiol. Cell Physiol. 279, C369–C374 (2000).
Louvard, D. et al. A monoclonal antibody to the heavy chain of clathrin. EMBO J. 2, 1655–1664 (1983).
Royle, S. J., Bright, N. A. & Lagnado, L. Clathrin is required for the function of the mitotic spindle. Nature 434, 1152–1157 (2005).
Hepler, P. K., McIntosh, J. R. & Cleland, S. Intermicrotubule bridges in mitotic spindle apparatus. J. Cell Biol. 45, 438–444 (1970).
Booth, D. G., Hood, F. E., Prior, I. A. & Royle, S. J. A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging. EMBO J. 30, 906–919 (2011).
Cheeseman, L. P., Harry, E. F., McAinsh, A. D., Prior, I. A. & Royle, S. J. Specific removal of TACC3/ch-TOG/clathrin at metaphase deregulates kinetochore fiber tension. J. Cell Sci. 126, 2102–2113 (2013).
Fu, W. et al. Clathrin recruits phosphorylated TACC3 to spindle poles for bipolar spindle assembly and chromosome alignment. J. Cell Sci. 123, 3645–3651 (2010).
Lin, C. H., Hu, C. K. & Shih, H. M. Clathrin heavy chain mediates TACC3 targeting to mitotic spindles to ensure spindle stability. J. Cell Biol. 189, 1097–1105 (2010).
Hubner, N. C. et al. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 189, 739–754 (2010).
Hood, F. E. et al. Coordination of adjacent domains mediates TACC3–ch-TOG–clathrin assembly and mitotic spindle binding. J. Cell Biol. http://dx.doi.org/10.1083/jcb.201211127 (2013).
Okamoto, M., Schoch, S. & Sudhof, T. C. EHSH1/intersectin, a protein that contains EH and SH3 domains and binds to dynamin and SNAP-25. A protein connection between exocytosis and endocytosis? J. Biol. Chem. 274, 18446–18454 (1999).
Pucharcos, C., Estivill, X. & de la Luna, S. Intersectin 2, a new multimodular protein involved in clathrin-mediated endocytosis. FEBS Lett. 478, 43–51 (2000).
Rodriguez-Fraticelli, A. E. et al. The Cdc42 GEF intersectin 2 controls mitotic spindle orientation to form the lumen during epithelial morphogenesis. J. Cell Biol. 189, 725–738 (2010).
Rosse, C., L'Hoste, S., Offner, N., Picard, A. & Camonis, J. RLIP, an effector of the Ral GTPases, is a platform for Cdk1 to phosphorylate epsin during the switch off of endocytosis in mitosis. J. Biol. Chem. 278, 30597–30604 (2003).
Fillatre, J. et al. Dynamics of the subcellular localization of RalBP1/RLIP through the cell cycle: the role of targeting signals and of protein–protein interactions. FASEB J. 26, 2164–2174 (2012).
Quaroni, A. & Paul, E. C. Cytocentrin is a Ral-binding protein involved in the assembly and function of the mitotic apparatus. J. Cell Sci. 112, 707–718 (1999).
Ma, M. P. & Chircop, M. SNX9, SNX18 and SNX33 are required for progression through and completion of mitosis. J. Cell Sci. 125, 4372–4382 (2012).
Hawryluk, M. J. et al. Epsin 1 is a polyubiquitin-selective clathrin-associated sorting protein. Traffic 7, 262–281 (2006).
Ford, M. G. et al. Curvature of clathrin-coated pits driven by epsin. Nature 419, 361–366 (2002).
Liu, Z. & Zheng, Y. A requirement for epsin in mitotic membrane and spindle organization. J. Cell Biol. 186, 473–480 (2009).
Greener, T., Zhao, X., Nojima, H., Eisenberg, E. & Greene, L. E. Role of cyclin G-associated kinase in uncoating clathrin-coated vesicles from non-neuronal cells. J. Biol. Chem. 275, 1365–1370 (2000).
Umeda, A., Meyerholz, A. & Ungewickell, E. Identification of the universal cofactor (auxilin 2) in clathrin coat dissociation. Eur. J. Cell Biol. 79, 336–342 (2000).
Shimizu, H., Nagamori, I., Yabuta, N. & Nojima, H. GAK, a regulator of clathrin-mediated membrane traffic, also controls centrosome integrity and chromosome congression. J. Cell Sci. 122, 3145–3152 (2009).
Tanenbaum, M. E. et al. Cyclin G-associated kinase promotes microtubule outgrowth from chromosomes during spindle assembly. Chromosoma 119, 415–424 (2010).
Borner, G. H. et al. Multivariate proteomic profiling identifies novel accessory proteins of coated vesicles. J. Cell Biol. 197, 141–160 (2012).
Foraker, A. B. et al. Clathrin promotes centrosome integrity in early mitosis through stabilization of centrosomal ch-TOG. J. Cell Biol. 198, 591–605 (2012).
Conner, S. D. & Schmid, S. L. Regulated portals of entry into the cell. Nature 422, 37–44 (2003).
Thompson, H. M., Cao, H., Chen, J., Euteneuer, U. & McNiven, M. A. Dynamin 2 binds γ-tubulin and participates in centrosome cohesion. Nature Cell Biol. 6, 335–342 (2004).
Tanabe, K. & Takei, K. Dynamic instability of microtubules requires dynamin 2 and is impaired in a Charcot–Marie–Tooth mutant. J. Cell Biol. 185, 939–948 (2009).
Thompson, H. M., Skop, A. R., Euteneuer, U., Meyer, B. J. & McNiven, M. A. The large GTPase dynamin associates with the spindle midzone and is required for cytokinesis. Curr. Biol. 12, 2111–2117 (2002).
Conery, A. R., Sever, S. & Harlow, E. Nucleoside diphosphate kinase Nm23-H1 regulates chromosomal stability by activating the GTPase dynamin during cytokinesis. Proc. Natl Acad. Sci. USA 107, 15461–15466 (2010).
Liu, Y. W., Surka, M. C., Schroeter, T., Lukiyanchuk, V. & Schmid, S. L. Isoform and splice-variant specific functions of dynamin-2 revealed by analysis of conditional knock-out cells. Mol. Biol. Cell 19, 5347–5359 (2008).
Konopka, C. A., Schleede, J. B., Skop, A. R. & Bednarek, S. Y. Dynamin and cytokinesis. Traffic 7, 239–247 (2006).
Joshi, S. et al. The dynamin inhibitors MiTMAB and OcTMAB induce cytokinesis failure and inhibit cell proliferation in human cancer cells. Mol. Cancer Ther. 9, 1995–2006 (2010).
Chircop, M. et al. Calcineurin activity is required for the completion of cytokinesis. Cell. Mol. Life Sci. 67, 3725–3737 (2010).
Gu, C. et al. Direct dynamin–actin interactions regulate the actin cytoskeleton. EMBO J. 29, 3593–3606 (2010).
Molla-Herman, A. et al. Targeting of β-arrestin2 to the centrosome and primary cilium: role in cell proliferation control. PLoS ONE 3, e3728 (2008).
Shankar, H. et al. Non-visual arrestins are constitutively associated with the centrosome and regulate centrosome function. J. Biol. Chem. 285, 8316–8329 (2010).
Mishra, S. K., Watkins, S. C. & Traub, L. M. The autosomal recessive hypercholesterolemia (ARH) protein interfaces directly with the clathrin-coat machinery. Proc. Natl Acad. Sci. USA 99, 16099–16104 (2002).
Mishra, S. K. et al. Functional dissection of an AP-2 β2 appendage-binding sequence within the autosomal recessive hypercholesterolemia protein. J. Biol. Chem. 280, 19270–19280 (2005).
Lehtonen, S. et al. The endocytic adaptor protein ARH associates with motor and centrosomal proteins and is involved in centrosome assembly and cytokinesis. Mol. Biol. Cell 19, 2949–2961 (2008).
Sun, X. M., Patel, D. D., Acosta, J. C., Gil, J. & Soutar, A. K. Premature senescence in cells from patients with autosomal recessive hypercholesterolemia (ARH): evidence for a role for ARH in mitosis. Arterioscler. Thromb. Vasc. Biol. 31, 2270–2277 (2011).
Traub, L. M. Tickets to ride: selecting cargo for clathrin-regulated internalization. Nature Rev. Mol. Cell Biol. 10, 583–596 (2009).
Smith, C. M. & Chircop, M. Clathrin-mediated endocytic proteins are involved in regulating mitotic progression and completion. Traffic 13, 1628–1641 (2012).
Young, J., Menetrey, J. & Goud, B. RAB6C is a retrogene that encodes a centrosomal protein involved in cell cycle progression. J. Mol. Biol. 397, 69–88 (2010).
Lara-Gonzalez, P., Westhorpe, F. G. & Taylor, S. S. The spindle assembly checkpoint. Curr. Biol. 22, R966–R980 (2012).
Kops, G. J. et al. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 169, 49–60 (2005).
Hirose, H. et al. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 23, 1267–1278 (2004).
Varma, D., Dujardin, D. L., Stehman, S. A. & Vallee, R. B. Role of the kinetochore/cell cycle checkpoint protein ZW10 in interphase cytoplasmic dynein function. J. Cell Biol. 172, 655–662 (2006).
Mallard, F. et al. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 156, 653–664 (2002).
Miserey-Lenkei, S. et al. A role for the Rab6A′ GTPase in the inactivation of the Mad2-spindle checkpoint. EMBO J. 25, 278–289 (2006).
Warren, G. Membrane partitioning during cell division. Annu. Rev. Biochem. 62, 323–348 (1993).
Fielding, A. B. & Royle, S. J. Mitotic inhibition of clathrin-mediated endocytosis. Cell. Mol. Life Sci. http://dx.doi.org/10.1007/s00018-012-1250-8 (2013).
Puhka, M., Vihinen, H., Joensuu, M. & Jokitalo, E. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J. Cell Biol. 179, 895–909 (2007).
Lucocq, J. M., Berger, E. G. & Warren, G. Mitotic Golgi fragments in HeLa cells and their role in the reassembly pathway. J. Cell Biol. 109, 463–474 (1989).
Tooze, J. & Hollinshead, M. Evidence that globular Golgi clusters in mitotic HeLa cells are clustered tubular endosomes. Eur. J. Cell Biol. 58, 228–242 (1992).
Waterman-Storer, C. M., Sanger, J. W. & Sanger, J. M. Dynamics of organelles in the mitotic spindles of living cells: membrane and microtubule interactions. Cell. Motil. Cytoskeleton 26, 19–39 (1993).
Niswonger, M. L. & O'Halloran, T. J. A novel role for clathrin in cytokinesis. Proc. Natl Acad. Sci. USA 94, 8575–8578 (1997).
Radulescu, A. E. & Shields, D. Clathrin is required for postmitotic Golgi reassembly. FASEB J. 26, 129–136 (2012).
Meraldi, P., Honda, R. & Nigg, E. A. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 21, 483–492 (2002).
Robinson, M. S., Sahlender, D. A. & Foster, S. D. Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria. Dev. Cell 18, 324–331 (2010).
Holland, A. J., Fachinetti, D., Han, J. S. & Cleveland, D. W. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc. Natl Acad. Sci. USA 109, E3350–E3357 (2012).
Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T. & Kanemaki, M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature Methods 6, 917–922 (2009).
Platani, M. et al. The Nup107–160 nucleoporin complex promotes mitotic events via control of the localization state of the chromosome passenger complex. Mol. Biol. Cell 20, 5260–5275 (2009).
Zuccolo, M. et al. The human Nup107–160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 26, 1853–1864 (2007).
Field, M. C. & Dacks, J. B. First and last ancestors: reconstructing evolution of the endomembrane system with ESCRTs, vesicle coat proteins, and nuclear pore complexes. Curr. Opin. Cell Biol. 21, 4–13 (2009).
Boettner, D. R., Chi, R. J. & Lemmon, S. K. Lessons from yeast for clathrin-mediated endocytosis. Nature Cell Biol. 14, 2–10 (2012).
Kakui, Y., Sato, M., Okada, N., Toda, T. & Yamamoto, M. Microtubules and Alp7–Alp14 (TACC–TOG) reposition chromosomes before meiotic segregation. Nature Cell Biol. 15, 786–796 (2013).
Scita, G. & Di Fiore, P. P. The endocytic matrix. Nature 463, 464–473 (2010).
Sigismund, S. et al. Endocytosis and signaling: cell logistics shape the eukaryotic cell plan. Physiol. Rev. 92, 273–366 (2012).
Drechsler, H. & McAinsh, A. D. Exotic mitotic mechanisms. Open Biol. 2, 120140 (2012).
Wilson, K. L. & Dawson, S. C. Evolution: functional evolution of nuclear structure. J. Cell Biol. 195, 171–181 (2011).
Gudise, S., Figueroa, R. A., Lindberg, R., Larsson, V. & Hallberg, E. Samp1 is functionally associated with the LINC complex and A-type lamina networks. J. Cell Sci. 124, 2077–2085 (2011).
Patel, H. et al. Kindlin-1 regulates mitotic spindle formation by interacting with integrins and Plk-1. Nature Commun. 4, 2056 (2013).
Kim, M. L., Sorg, I. & Arrieumerlou, C. Endocytosis-independent function of clathrin heavy chain in the control of basal NF-κB activation. PLoS ONE 6, e17158 (2011).
Enari, M., Ohmori, K., Kitabayashi, I. & Taya, Y. Requirement of clathrin heavy chain for p53-mediated transcription. Genes Dev. 20, 1087–1099 (2006).
Ybe, J. A. et al. Nuclear localization of clathrin involves a labile helix outside the trimerization domain. FEBS Lett. 587, 142–149 (2013).
Hupalowska, A. & Miaczynska, M. The new faces of endocytosis in signaling. Traffic 13, 9–18 (2012).
Vecchi, M. et al. Nucleocytoplasmic shuttling of endocytic proteins. J. Cell Biol. 153, 1511–1517 (2001).
Fielding, A. B., Willox, A. K., Okeke, E. & Royle, S. J. Clathrin-mediated endocytosis is inhibited during mitosis. Proc. Natl Acad. Sci. USA 109, 6572–6577 (2012).
Schweitzer, J. K., Burke, E. E., Goodson, H. V. & D'Souza-Schorey, C. Endocytosis resumes during late mitosis and is required for cytokinesis. J. Biol. Chem. 280, 41628–41635 (2005).
Belgareh, N. et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 154, 1147–1160 (2001).
Doxsey, S. J., Brodsky, F. M., Blank, G. S. & Helenius, A. Inhibition of endocytosis by anti-clathrin antibodies. Cell 50, 453–463 (1987).
Goud, B., Huet, C. & Louvard, D. Assembled and unassembled pools of clathrin: a quantitative study using an enzyme immunoassay. J. Cell Biol. 100, 521–527 (1985).
Brodsky, F. M. Diversity of clathrin function: new tricks for an old protein. Annu. Rev. Cell Dev. Biol. 28, 309–336 (2012).
Taylor, M. J., Perrais, D. & Merrifield, C. J. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 9, e1000604 (2011).
Brodsky, F. M., Chen, C. Y., Knuehl, C., Towler, M. C. & Wakeham, D. E. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu. Rev. Cell Dev. Biol. 17, 517–568 (2001).
Chircop, M. et al. Phosphorylation of dynamin II at serine-764 is associated with cytokinesis. Biochim. Biophys. Acta 1813, 1689–1699 (2011).
Lee, D. W., Zhao, X., Yim, Y. I., Eisenberg, E. & Greene, L. E. Essential role of cyclin-G-associated kinase (auxilin-2) in developing and mature mice. Mol. Biol. Cell 19, 2766–2776 (2008).
Acknowledgements
The author thanks L. Wood for useful comments and his colleagues in Liverpool and at Warwick Medical School for interesting discussions on moonlighting functions. He is supported by a Senior Cancer Research Fellowship from Cancer Research UK (C25425/A15182) and a project grant from the UK Biotechnology and Biological Sciences Research Council (BBSRC) (BB/H015582/1).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Rights and permissions
About this article
Cite this article
Royle, S. Protein adaptation: mitotic functions for membrane trafficking proteins. Nat Rev Mol Cell Biol 14, 592–599 (2013). https://doi.org/10.1038/nrm3641
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3641
This article is cited by
-
GRP75-driven, cell-cycle-dependent macropinocytosis of Tat/pDNA-Ca2+ nanoparticles underlies distinct gene therapy effect in ovarian cancer
Journal of Nanobiotechnology (2022)
-
Whole-genome resequencing reveals genetic indels of feathered-leg traits in domestic chickens
Journal of Genetics (2019)
-
Automated classification and characterization of the mitotic spindle following knockdown of a mitosis-related protein
BMC Bioinformatics (2017)
-
RalBP1 and p19-VHL play an oncogenic role, and p30-VHL plays a tumor suppressor role during the blebbishield emergency program
Cell Death Discovery (2017)
-
K-Cl cotransporter KCC2—a moonlighting protein in excitatory and inhibitory synapse development and function
Pflügers Archiv - European Journal of Physiology (2015)