Detecting trace quantities of substances in complex aqueous media, such as river water, sewage and treated drinking water, can be like looking for the proverbial needle in a haystack. Numerous sensitive methods have been developed1, but they often rely on complicated sample-processing and signal-acquisition steps, and are typically time consuming and costly. An approach known as surface-enhanced Raman spectroscopy (SERS) can detect single molecules2,3 and has long been promoted as a promising alternative to such methods. But SERS is a notoriously finicky technique, with intrinsic variabilities, which makes it hard to use to quantify low concentrations of dissolved compounds. Writing in Nature, Bi et al.4 report a digitized SERS method for detecting waterborne substances that addresses this fundamental issue.
Read the paper: Digital colloid-enhanced Raman spectroscopy by single-molecule counting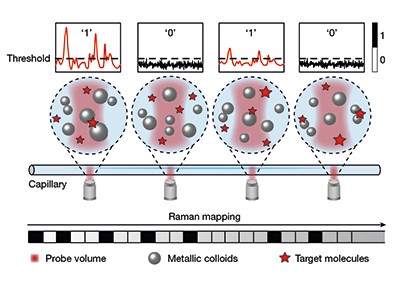
Discovered in 1928, Raman spectroscopy is a laser-based technique that depends on the inelastic scattering of light by molecules — a phenomenon in which light excites a vibration in the molecules, producing scattered photons that have a different energy than do the incoming photons. The frequencies of the scattered photons depend on the bonds in the molecules, and thus provide a diagnostic fingerprint that readily differentiates one type of molecule from another. Unfortunately, the signal intensities in Raman spectroscopy are inherently weak and therefore challenging to detect. This issue has long hindered the use of Raman spectroscopy for practical applications.
The outlook for Raman-based diagnostic techniques changed, however, after the unexpected discovery in 1974 that the Raman scattering signal can be greatly increased when a molecule is adsorbed to a roughened silver electrode5. Subsequent studies6 have shown that the signal enhancement arises as a result of interactions between the molecule and electrons on the metal surface. This enhancement forms the basis of the SERS technique.
A key challenge that undermines the analytical capability of SERS is the spatial and temporal variability of the signal enhancement. This variation arises because a combination of events is required to detect molecules in solution. First, a dissolved molecule must travel to the surface of a metal (typically gold or silver) and adsorb to that surface. Crucially, to achieve the lowest detection limits, the molecule must adsorb at a SERS ‘hotspot’ — a region of the surface with geometric and electronic characteristics that best enhance the Raman signal — and it must then reside there for long enough to facilitate signal enhancement. Given that molecular motion and adsorption are random processes, and that hotspots can be challenging to fabricate reproducibly, it has been difficult to develop SERS methods that consistently and reliably quantify the concentration of molecules in solution.
Squeezed light improves sensitivity of microscopy technique
Improvements in reproducibility and quantification have been achieved by precise engineering of metal surfaces6 and through hotspot normalization7 — a process that calibrates the intensity of Raman signals on the metal surface by using inelastic scattering of photons at the surface as an internal reference. Digital SERS methods have also been reported8 in which the number of spatially discrete Raman signals is counted, rather than signal intensities being measured — mostly as a way to improve the detection limits of surface-bound arrays of metal nanoparticles. Although such arrays are valuable for many applications, they cannot be used in all circumstances (in biological cells, for example).
Bi et al. now report a digital SERS protocol that uses colloids — nanoparticles dispersed in aqueous solution — rather than surface-bound nanoparticles. In this approach (Fig. 1), the authors mix silver colloid particles (20 to 50 nanometres in diameter) with target molecules to produce colloidally stable (minimally aggregated), uniform suspensions. SERS spectra are acquired at thousands of positions (voxels) in a capillary tube filled with the suspension, and are then digitized by considering the signal intensities measured at each position: signals exceeding a chosen threshold value correspond to the detection of a single target molecule and are given a value of 1, whereas those below the threshold correspond to no detection and are assigned a value of 0.
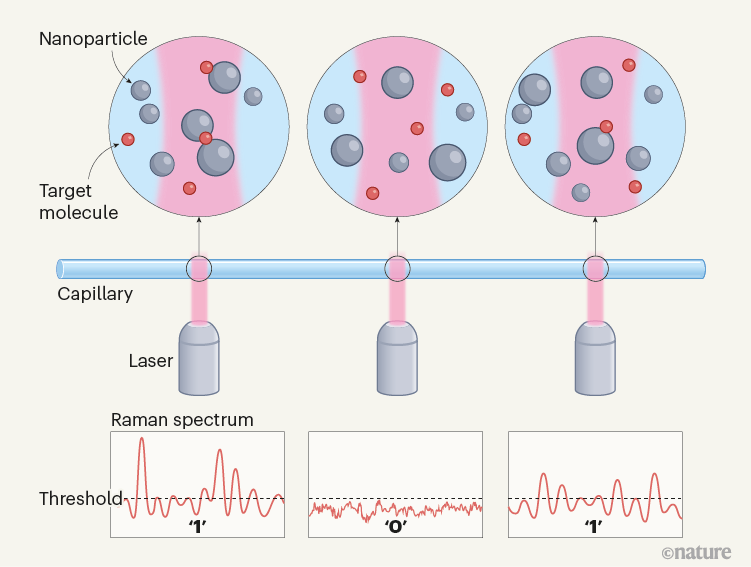
Figure 1 | Digitization of surface-enhanced Raman spectroscopy (SERS) using suspended nanoparticles. Bi et al.4 report a technique for quantifying ultralow concentrations of dissolved molecules in water. A suspension of silver nanoparticles is mixed with a solution of the target molecules, placed in a capillary tube and then irradiated by light at sequential positions along the tube. When target molecules adsorb to the surface of a nanoparticle, inelastic light scattering sometimes occurs, producing a signal known as a Raman spectrum. The signal is assigned a value of 1 when the amplitude of the spectral peaks exceeds a predetermined threshold, indicating that a target molecule has been detected. When the amplitude does not exceed the threshold, the signal is given a value of 0, indicating no detection. The total number of spatially discrete detections in the experiment is used to determine the concentration of target molecules. (Adapted from Fig. 1a of ref. 4.)
Because the number of detection events per unit volume follows a Poisson statistical distribution, the total number of events measured in a sample reflects the concentration of target molecules. This approach gives reproducible measurements of ultralow concentrations (down to 10–13 molar), and overcomes the problems of signal variability by measuring the number of detection signals, rather than their intensity.
Bi and colleagues’ extension of the digital SERS approach to suspended colloidal particles greatly expands the portfolio of conditions under which the technique can be used for molecular detection. With their method, a sample suspected to contain a target molecule can be collected, processed and analysed using silver nanoparticles, which are inexpensive and easy to produce. For example, the authors show that they can detect toxic herbicides and fungicides in lake water and in extracts of bean sprouts.
An in situ search for organic molecules in Mars’ Jezero Crater
A limitation of the study is that the target molecules are restricted to those that readily associate with the surface of a noble metal (such as silver). The authors use several surface-engineering approaches to strengthen the interactions between the targets and the metal, but such enhancement might be challenging for all molecules of interest. Furthermore, the method as it stands involves a potentially cumbersome calibration, in which optimal nanoparticle concentrations and signal-acquisition times need to be determined for each target molecule before that molecule can be analysed. Given that future applications will involve diverse targets and increasingly large data sets, it will be interesting to investigate whether machine-learning methods could be used to develop general rules that relate the structures of target molecules to appropriate conditions for analysis.
Finally, the samples being analysed currently require extensive dilution to attenuate any possible signal interference from non-target substances in the aqueous medium, and the extent of dilution needs to be experimentally determined for each system. Such an approach will not always be practical, and will often prolong the time between sample acquisition and quantification.
It is likely that these issues will be resolved as the new method is put to the test by analytical chemists and forensic scientists. In the meantime, Bi et al. have gifted researchers working with SERS a fitting 50th birthday present, which will help to push this exciting method towards true field applications.

 Read the paper: Digital colloid-enhanced Raman spectroscopy by single-molecule counting
Read the paper: Digital colloid-enhanced Raman spectroscopy by single-molecule counting
 Squeezed light improves sensitivity of microscopy technique
Squeezed light improves sensitivity of microscopy technique
 An in situ search for organic molecules in Mars’ Jezero Crater
An in situ search for organic molecules in Mars’ Jezero Crater