Abstract
The present prospective cohort study evaluated the prevalence of FSH-R receptor Asn680Ser and Ala307Thr among infertile Indian women and the correlation of these polymorphisms with ART outcomes. Total 804 infertile and 209 fertile controls were enrolled for FSH-R analysis. Correlation of different genotypes with ovarian reserve markers, IVF parameters, and cumulative live birth rates (CLBR) was done among women undergoing IVF. In fertile controls, at 680 position GG (Ser/Ser) was the most common genotype; but among infertile women, all the genotypes were equally distributed. There was no significant difference in ovarian response parameters, oocyte yield, and CLBR among the three genotype groups. Empty follicle syndrome (EFS) was highest in women with AA or AG type at both positions. On categorisation of unexpected poor responders according to POSEIDON stratification; GG genotype at both positions had the lowest risk ratio of low-oocyte yield in ART cycles, but these differences were not statistically significant. This is the largest study from Indian ethnicity showing GG (Ser/Ser) genotype is most common among fertile women. The effect of FSH-R genotypes is very marginal on IVF parameters and is not reflected in CLBR. More prospective data may be required on the correlation of these genotypes with genuine EFS, thus stratifying the next cycles with self or donor oocytes. Routine genetic testing of FSH-R polymorphism should not be done except in a research setting. As both 680 and 307 positions are in linkage disequilibrium, only 680 position analysis may be done in a research setting.
Similar content being viewed by others
Introduction
Assessment of ovarian reserve markers helps the clinician with tailored and individualised ovarian stimulation protocol for infertile women undergoing in vitro-fertilisation (IVF) procedures [1]. Parameters like chronological age, ovarian reserve markers (baseline serum follicle stimulating hormone (FSH) and anti-mullerian hormone (AMH), BMI, ovarian response in previous stimulation cycles, or combination of these factors are used for starting gonadotrophin dose calculation for each woman. However accurate prediction of the ovarian response to exogenous FSH stimulation is not yet possible and unpredictability of ovarian response among different women has triggered the research in pharmacogenetic pathways modulating the ovarian response to FSH injections [2]. Discovering the genetic variants associated with the ovarian response to gonadotrophins may be an important step towards individualised protocols for ovarian stimulation [3]. Pharmacogenomic approach describes ‘the individualisation of treatment based on a patient’s genetic profile’, which may prove to be a novel patient-tailored approach in women undergoing IVF cycles.
Since last two decades, research is being focussed on identification of biomarkers for FSH and its receptors in ovarian granulosa cells which modulate the ovarian response to exogenous gonadotrophins in ART. Some mutations, such as Ile160Thr, Ala189Val, and Asn191Ile, are associated with complete inhibition of FSH-R activity [4]. Other single nucleotide polymorphisms (SNPs) in the FSH-R gene lead to either excessive gonadotrophin consumption, increased duration of stimulation, and can influence the number of mature oocytes retrieved in IVF [5, 6] or can increase the chances of hyper-response, thus increasing chances of ovarian hyperstimulation [7].
Most commonly studied SNPs among these include Thr307Ala (rs6165) and Asn680Ser (rs6166) [8, 9]. These two single nucleotide polymorphisms (SNPs) T307A (rs6165) and N680S (rs6166), are in complete linkage disequilibrium and do not show random distribution [5]. Same linkage disequilibrium of these two SNPs has been reported by Simoni and Casarini except in the African population [10].
Considering the fact that these polymorphisms may lead to partial interference in FSH receptor function, a better understanding of these may prove useful tool for predicting ovarian response in women undergoing IVF cycles. Although, Ser/ser (GG) genotype at position 680 has been reported to cause more FSH consumption and less oocyte yield, the effect of these polymorphisms on live birth has not been found significant [11,12,13]. The results are also inconsistent due to heterogenicity in inclusion criteria, including women with variable ovarian reserve parameters, different gonadotrophin doses, and allowing dose adjustments during treatments [9]. There is still a need to have more data on ethnic distribution of these SNPs, their actual association with ovarian response, and their role in reducing the overall cost of IVF treatment in unexpectedly poor responders going for repeated IVF cycles.
The present prospective study evaluated the prevalence of FSH-R polymorphism of Asn680Ser (rs6166) and Thr307Ala (rs6165)) in infertile cases and fertile controls. The study also evaluated the correlation of these polymorphisms with ovarian reserve markers, association with ovarian response, and impact on IVF/ICSI outcomes in Indian women.
Methodology
This was a prospective cohort study conducted over a period of three and half years (October 2018–March 2022). The case group consisted of 804 sub-fertile women consulting in the infertility clinic of a tertiary care referral hospital. The control group included 209 fertile controls; which were recruited from voluntary oocyte donors and fertile women visiting gynaecology clinic. The screening was started after institute ethical approval (IEC -515/ 01.09.2017). and recruitment was done after informed written consent.
Inclusion criteria for infertile cases were: age between 22 and 38 years; BMI 19 to 30 kg/m2; tubal, male, or unexplained factor infertility. Exclusion criteria were: endometriosis; history of ovarian surgery; polycystic ovarian syndrome; uncontrolled endocrine disorder (thyroid disorder or diabetes); baseline serum FSH > 12 mIU/ml and AMH < 1 ng/ml.
Study procedure
Detailed history of infertility including previous infertility treatment, IVF details including response to stimulation and outcome (if available) was recorded, and a clinical examination was done. Serum samples for FSH, LH, AMH, prolactin, and thyroid-stimulating hormone (TSH) were taken. Transvaginal scan for antral follicle count and uterine cavity evaluation was done. Five ml of blood sample were taken from the study participants and stored in sterile EDTA vacutainers and stored at −80 °C for later use. DNA extraction was done, and FSH-R analysis.
Controlled ovarian hyperstimulation
Of the total study participants, 423 women in the case group and 46 voluntary oocyte donors from the control group underwent ART. The IVF protocols used were either antagonist or agonist protocols. The decision for selection of protocol and starting dose of the gonadotrophin was left for the clinician in-charge to decide depending upon patient age; BMI, and previous ovarian response if available.
Follicle monitoring was done starting from day 6 of stimulation onwards. Recombinant human chorionic gonadotropin (hCG, Ovitrelle; Merck Serono, Italy) 250 μg s.c was administered for inducing ovulation when at least 3 follicles were more than 18 mm in size. Oocyte retrieval was performed 34–36 h after hCG administration and IVF/ICSI was done as per embryologist discretion.
Embryos transfer was done at at cleavage (Day 2 or 3) or blastocyst stage (day5) depending on the patient profile and status of embryo number and quality. Micronized progesterone 100 mg intramuscularly or vaginal progesterone 600 mg/day was given after ET. Clinical pregnancy was defined as viable gestation at 6 weeks scan and ongoing pregnancy as viable gestation beyond 12 weeks. The cumulative live birth rate (CLBR) per aspiration cycle was calculated with numerator as number of women having at least one live birth, and denominator was taken as all the women with either at least one live birth and those where all the embryos were exhausted. Further, the cohort who underwent IVF was segregated into normo-responders and different POSEIDON groups according to the number of oocyte retrieved. Analysis was done to evaluate the correlation of FSH-R polymorphisms among different POSEIDON groups. CLBR analysis was not done for controls (N = 46) as they were all fertile oocyte donors.
Statistical analysis
Statistical analysis was carried out using Stata 15.0 (StataCorp LLP, Texas, USA). Continuous variables were summarised as mean ± SD/median (P25 – P75) as appropriate, and categorical variables were summarised as number and percentage. The distribution of A and G allelles of SNP 680 and SNP 307 (three possible genotypes) was assessed using the Hardy–Weinberg Equilibrium and tested using chi-square test for goodness-of-fit with one degrees of freedom among cases and controls. Demographic variables were compared between cases and controls women using unpaired ‘t’ test. The association of genotype with ovarian response variables, baseline hormonal parameters, fertilisation, and cleavage rates among sub-fertile women was tested using one-way ANOVA or Kruskal–Wallis test. The normality of continuous variables were checked using Shapiro–Wilks test. The odds ratio and 95% confidence interval were calculated to assess the effect of genotypes between cases and controls. The association of genotypes with POSEIDON (defined using age and oocyte yield) stratification and CLBR among cases who underwent IVF was analysed using multinomial logistic regression, and results are reported as a relative-risk ratio along with 95% CI. The p value <0.05 was considered statistically significant.
Results
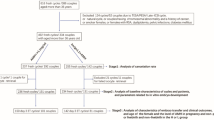
Figure 1 describes the flow diagram of the study. The baseline characteristics of the sub-fertile patients and fertile control group is as shown in Table 1. Although, there was significant statistical difference in mean age, BMI, AFC (antral follicle count), serum FSH and serum AMH levels between study and control groups, but it was not significant clinically.
As shown in Table 2, our ethnic population followed the Hardy-Weinberg equilibrium for SNP 307 in cases only (p > 0.05), and rest were in linkage disequilibrium (p < 0.05).
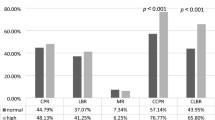
The distribution of allele ‘A’ was higher among cases at both SNP 680 & 307 while ‘G’ allele frequency was higher among cases at SNP 680 and comparable in both groups at SNP 307. Looking at genotype frequency distribution; GG genotype was significantly higher in fertile controls (56.5%) as compared to infertile cases. In infertile cohort, although all the three genotypes were equally distributed, but AA and AG were significantly higher in infertile cohort as compared to fertile control (P < 0.01). At position 307, the most common genotype was the heterozygous in both fertile and infertile women, infertile women had significantly higher genotypes with allele “A”.
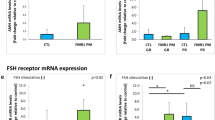
Serum FSH was significantly lower in GG genotype at both 680 and 307 positions. At position 307, AMH was also significantly higher in GG subtype (Table 3).
There was no significant difference in ovarian response parameters like duration of stimulation, dose of gonadotropins, follicle number on the day of trigger, serum E2 levels on the day of trigger, number of retrieved oocytes, grade I COCs (cumulus-oocyte complexes with well-expanded cumulus, radiant corona, distinct zona pellucida, and clear ooplasm) [14] and CLBR in between the three genotypes at both SNP 680 and SNP 307 except for total dose requirement significantly higher in GG genotype and cleavage rate highest in heterozygous sub-group (Table 4). Out of 423 aspiration cycles done in infertile group, embryo transfer (ET) was done in 383 cycles; 40 cycles didn’t undergo ET secondary to empty follicle syndrome (EFS), fertilisation failure, cleavage failure, or freeze-all. Clinical pregnancy rate (CPR) per embryo transfer was 26% (121 pregnancies out of 464 transfers done).On further distribution; CCPR in fresh ET was 20.3% (62/306), FET I was 38.4% (43/112), FET II was 37.1% (13/35), and FET III (3/11) was 27.3%. So total failed embryo transfers was 343.
Incidence of genuine EFS was 3.3% among cases 4.3% among controls. Most of the genuine EFS belonged to AA and AG genotypes at both positions and both in cases and controls. In women with serum estrtadiol <1000 pg/ml, the maximum belonged to genotype AA followed by AG and the least with GG genotype at both SNP 680 and 370 positions (Supplementary Fig. s1).
The whole cohort who underwent IVF (N = 423) was subdivided into normo-responders and unexpected poor responders according to POSEIDON (female age and AMH levels). GG genotype was least in POSEIDON Ia group at both 680 and 307 positions. The most commonly seen genotype among all POSEIDON sub-group was heterozygous subtype at both positions (Fig. 2). However, these distribution differences were not statistically significant. The relative-risk ratio and 95% CI shown for various categories of POSEIDON as well as CLBR with SNP 680 and SNP 307 was not statistically significant (Fig. 3a, b).
Correlation of POSEIDON stratification and cumulative live birth rate (CLBR) with SNP 680 and SNP 307. a Forest-plot showing the relative-risk ratio (RRR) (95% CI) of unexpected poor responders (POSEIDON I & II) with genotype at position 680 and 307 of FSH-R. b Forest-plot showing relative-risk ratio (RRR) (95% CI) of cumulative live birth rate (CLBR) in POSEIDON I & II with genotype at position 680 and 307 of FSH-R
Discussion
The present prospective study evaluated the prevalence of FSH-R receptor polymorphisms among infertile and fertile women of Indian-Asian ethnicity. The study concluded that GG (Ser/Ser) is the most common genotypes among fertile Indian women at SNP 680. The genotypes did not significantly affect the ART outcomes on sub-group analysis of infertile cases into normo-responders and unexpected poor-responders at both SNPs.
The first study from India on FSH- R genotypes was done by Achrekar et al. [8] in 2009, which showed heterozygous genotype were most common at both 680 and 307 position among fertile and infertile women. Also, the distribution of different sub-types was comparable among cases and controls. In our study, GG (Ser/Ser) at position 680 was the most common genotype among fertile controls. Rest distribution of all genotypes at both positions showed heterozygous genotype (Asn/Ser (AG) at 680; Ala/Thr (AG) at 307) to be most common with infertile women having significantly higher genotypes with allele “A”(p < 0.01). Previous studies have reported similar observations [13, 15].
The interesting genotype; Asn/Ser at position 680 has been studied extensively, and different conclusions have been reported in different studies from different ethnic populations. Latest Delphi consensus statement by Conforti A et al. [16] and previous studies [17,18,19,20] have reported that Ser/Ser (position 680) is associated with higher basal FSH levels; higher requirement of gonadotrophins and less number of oocytes. Similarly in report by Nenonen et al. [7], the relative risk of development of ovarian hyperstimulation syndrome (OHSS) was significantly higher in women with SNP AA and AG at position 680 as compared to those with GG. Recently, Baldini et al. studied the effect of FSH-R 680 on ovarian response in ART cycles and further divided them into abnormal and normal responders. He concluded that in abnormal responders, a significant prevalence of the amino acid serine at position 680 was detected (p < 0.05) [20]. Similar to the above study; Bayraktar et al. [21] also observed more number of poor-responders in Ser/Ser group among Turkish women.
There are two studies published till date which are in concordance with our study. First study which reported the findings against Perez Mayorga et al. was by Klinkert et al. who studied SNP 680 and its relation with IVF response and IVF outcomes. The ovarian response was comparable between patients with different FSH receptor genotypes, and patients with polymorphism Ser/Ser (GG) had implantation and pregnancy rates three times higher compared with patients with polymorphism Asn/Asn [22].
A previous study on Indian ethnicity by Achrekar et al. evaluated the combined effect of the polymorphisms at positions −29 and 680 of FSH-R with the type of ovarian response and receptor expression. Various clinical parameters revealed that 75% of the subjects with A/A–Asn/Asn (AA) genotype were poor ovarian responders (odds ratio 7.92; P = 0.009). The study speculated that the A allele at position −29 and the Asn allele at position 680 (AA) might be more susceptible to poor ovarian response. Although our study evaluated only SNP 680 polymorphism, we have reported that Ser/Ser (GG) at 680 is most common among fertile controls, and least risk of poor oocyte yield and least association with EFS. Despite this, it did not affect the CLBR, which was comparable among all the genotypes.
EFS is one unpredictable outcome in IVF cycles with wide range of prevalence reported upto of 0.045–7% of aspirated IVF cycles [23]. Lazaros et al. reported that lower serum FSH levels, higher follicle and oocyte numbers, increased numbers of large follicles as well as decreased empty follicle numbers in Thr307Thr/Asn680Asn (AA) women as compared to Thr307 Ala/Asn680Ser (AG) and Ala307Ala/Ser680Ser (GG) women (p < 0.006, p < 0.01, p < 0.008, p < 0.01, p < 0.005, respectively). Contradictory to this study, most of the patients in our cohort with EFS despite high serum hCG levels, had either AA or AG genotype both at 680 and 307 positions; implying that presence of ‘A’ allele in Indian-Asian women may cause resistance to FSH action. Also lowest peak oestradiol levels (<1000 pg/ml) were observed among AA genotype at both SNP 680 and 307.
Despite these differences, most of the studies have reported comparable IVF outcomes among different genotype subgroups. Jun et al. [24] found highest pregnancy rate per transfer with AA genotype and the lowest with GG genotype at SNP 680. While Klinkert et al. [22] reported that Ser680Ser was associated with higher pregnancy rates and implantation rates. Konig et al. [13] reported comparable CLBR among different genotypes despite Ser/Ser having higher basal FSH levels and a significantly lower number of oocytes and embryos. Lindgren et al. [25] found that women homozygous for Ser at both FSH-R and LHR had ~40% higher chance of live birth in the first IVF cycle with a doubled chance in cumulative cycles. CLBR in the present study although numerically higher in the heterozygous group (Asn/Ser and Ala/Thr), but the difference was not statistically significant among three genotypes similar to those published by Konig et al. [13].
Studies have evaluated the effect of 307 SNP polymorphism and reported Ala/Ala (GG) genotype to be significantly associated with the use of higher doses of recombinant FSH; suggesting lower sensitivity of ‘G’ allele to r-FSH but CLBR was comparable among three genotypes, at 307 position [5].
The strengths of our study are the largest study cohort of Indian ethnicity along with fertile control group where we had IVF details available for 46 controls (voluntary oocyte donors). This is the first study to divide the IVF cohort into normal-responders and unexpected poor responders and POSEIDON I and II stratification according to IVF response. This is the first study from India to stratify all unexpected poor-responders according to POSEIDON stratification and correlation of different genotypes with different POSEIDON groups. Patients with GG genotype showed the lowest risk of low-oocyte-yield in IVF/ICSI cycles at both SNPs. The results are similar to previous studies by Achrekar et al. and Klinkert et al. and not in concordance with other reports and meta-analyses. This is the first study elaborating the EFS cases and their relation to polymorphisms and was found maximum in women with allele A at both positions.
Limitations are that not all the women recruited could undergo IVF during COVID pandemic. COVID also affected oocyte donor recruitments. Dose selection and adjustments were according to clinician discretion which may have affected oocyte yield. Although our centre is tertiary referral centre, future prospective studies with different centres in the country may give more generalisable information.
To conclude; GG genotype in Indian-Asian women (Ser/Ser 680 and Ala/Ala 307) is characterised by lower basal FSH levels and is more common among fertile women. The effects of these polymorphisms on IVF parameters are small and negligible, with no significant effect on oocyte yield and CLBR. The presence of allele “A” (asparagine and threonine) may be responsible for genuine EFS and unexpected low estradiol levels in IVF cycles in Indian women. The results pave the way for new studies based on pharmacogenomics, randomised controlled prospective, and multi‐centre studies from different geographical areas before utilising FSH-R polymorphism as a confounder for the deficient ovarian response in ART.
References
Sunkara SK, Rittenberg V, Raine-Fenning N, Bhattacharya S, Zamora J, Coomarasamy A. Association between the number of eggs and live birth in IVF treatment: an analysis of 400 135 treatment cycles. Hum Reprod. 2011;26:1768–74.
Altmäe S, Hovatta O, Stavreus-Evers A, Salumets A. Genetic predictors of controlled ovarian hyperstimulation: where do we stand today? Hum Reprod Update. 2011;17:813–28.
Casarini L, Simoni M. Gene polymorphisms in female reproduction. Methods Mol Biol. 2014;1154:75–90.
Dieamant F, Petersen CG, Vagnini LD, Petersen B, Ricci J, Nicoletti A, et al. The Ala307Thr polymorphism of the follicle-stimulating hormone receptor (FSHR) gene is associated with the dose of recombinant FSH received during IVF/ICSI treatment. JBRA Assist Reprod. 2023;27:78–84.
Perez Mayorga M, Gromoll J, Behre HM, Gassner C, Nieschlag E, Simoni M. Ovarian response to follicle-stimulating hormone (FSH) stimulation depends on the FSH receptor genotype. J Clin Endocrinol Metab. 2000;85:3365–9.
Alviggi C, Conforti A, Santi D, Esteves SC, Andersen CY, Humaidan P, et al. Clinical relevance of genetic variants of gonadotrophins and their receptors in controlled. ovarian stimulation: a systematic review and meta-analysis. Hum Reprod Update. 2018b;24:1–16.
Nenonen HA, Lindgren IA, Prahl AS, Trzybulska D, Kharraziha I, Hultén M. The N680S variant in the follicle-stimulating hormone receptor gene identifies hyperresponders to controlled ovarian stimulation. Pharmacogenet Genomics 2019;29:114–20.
Achrekar SK, Modi DN, Desai SK, Mangoli VS, Mangoli RV, Mahale SD. Follicle-stimulating hormone receptor polymorphism (Thr307Ala) is associated with variable ovarian response and ovarian hyperstimulation syndrome in Indian women. Fertil Steril. 2009;91:432–9.
Plozos NP, Neves AR, Drakopoulos P, Spits C, Mercadal BA, Gracia S, et al. The effect of polymorphisms in FSHR and FSHB genes on ovarian response: a prospective multicenter multinational study in Europe and Asia. Hum Reprod. 2021;36:1711–21.
Simoni M, Casarini L. Mechanisms in endocrinology: genetics of FSH action a 2014-and-beyond view. Eur J Endocrinol. 2014;170:R91–107.
La Marca A, Sighinolfi G, Argento C, Grisendi V, Casarini L, Volpe A, et al. Polymorphisms in gonadotropin and gonadotropin receptor genes as markers of ovarian reserve and response in. in vitro fertilization. Fertil Steril. 2013;99:970–8.e1.
Trevisan CM, Peluso C, Cordts EB, R De O, Christofolini DM, Barbosa CP, et al. Ala307Thr and Asn680Ser polymorphisms of FSHR gene in human reproduction outcomes. Cell Physiol Biochem. 2014;34:1527–35.
König TE, J van der L, Schats R, Lambalk CB. The relationship between FSH receptor polymorphism status and IVF cycle outcome: a retro- spective observational study. Reprod Biomed Online. 2019;39:231–40.
Lin YC, Chang SY, Lan KC, Huang HW, Chang CY, Tsai MY, et al. Human oocyte maturity in vivo determines the outcome of blastocyst development in vitro. J Assist Reprod Genet. 2003;20:506–12.
Kuijper EAM, Blankenstein MA, Luttikhof LJ, Roek SJM, Overbeek A, Hompes PG, et al. Frequency distribution of polymorphisms in the FSH receptor gene in infertility patients of different ethnicity. Reprod Biomed Online. 2011;22:S60–65.
Conforti A, Tüttelmann F, Alviggi C, Behre HM, Fischer R, Hu L, et al. Effect of genetic variants of gonadotropins and their receptors on ovarian stimulation outcomes: a delphi consensus. Front Endocrinol (Lausanne). 2022;12:797365.
Yan Y, Gong Z, Zhang L, Li Y, Li X, Zhu L, et al. Association of follicle-stimulating hormone receptor polymorphisms with ovarian response in Chinese women: a prospective clinical study. PloS One. 2013;8:e78138.
Huang X, Li L, Hong L, Zhou W, Shi H, Zhang H, et al. The Ser680Asn polymorphism in the follicle-stimulating hormone receptor gene is associated with the ovarian response in controlled ovarian hyperstimulation. Clin Endocrinol. 2015;82:577–83.
Desai SS, Achrekar SK, Paranjape SR, Desai SK, Mangoli VS, Mahale SD. Association of allelic combinations of FSHR gene polymorphisms with ovarian response. Reprod Biomed Online. 2013;27:400–6.
Baldini MG, Catino A, Palini S, Sciorio R, Ferri D, Vinciguerra M, et al. The polymorphism Asn680Ser on the FSH receptor and abnormal ovarian response in patients with normal values of AMH and AFC. Int J Mol Sci 2023;24:1080.
Bayraktar B, Güleç ES, Kutbay YB, Köse C, Gür EB, Demir A. Does follicle-stimulating hormone receptor polymorphism status affect In vitro fertilization-intracytoplasmic sperm injection results and live birth rate? A retrospective study. J Hum Reprod Sci. 2022;15:58–63.
Klinkert ER, te Velde ER, Weima S, van Zandvoort PM. Hanssen RGJM, Nilsson PR, et al. FSH receptor genotype is associated with pregnancy but not with ovarian response in IVF. Reprod Biomed Online. 2006;13:687–95.
Revelli A, Carosso A, Grassi G, Gennarelli G. Stefano Canosa 2, Benedetto C. Empty follicle syndrome revisited: definition, incidence, aetiology, early diagnosis and treatment. Reprod Biomed Online. 2017;35:132–8.
Jun JK, Yoon JS, Ku S-Y, Choi YM, Hwang KR, Park SY, et al. Follicle-stimulating hormone receptor gene polymorphism and ovarian responses to controlled ovarian hyperstimulation for IVF-ET. J Hum Genet. 2006;51:665–70.
Lindgren I, Nenonen H, Henic E, Bungum L, Prahl A, Bungum M, et al. Gonadotropin receptor variants are linked to cumulative live birth rate after in vitro fertilization. J Assist Reprod Genet. 2019;36:29–38.
Funding
The study was done with funding from Science and Engineering Research Board (SERB), as an extramural funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
The study was done after ethical approval from the Institute Ethics Committee (IEC).
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mahey, R., Rajput, M., Dada, R. et al. Prevalence of FSH-R Asn680Ser and Ala307Thr receptor polymorphism and their correlation with ART outcomes among infertile Indian-Asian women-a prospective cohort study. J Hum Genet (2024). https://doi.org/10.1038/s10038-024-01251-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s10038-024-01251-8