Abstract
Mucopolysaccharidosis (MPS) type II is caused by a deficiency of iduronate-2-sulfatase and is characterized by the accumulation of glycosaminoglycans (GAGs). Without effective therapy, the severe form of MPS II causes progressive neurodegeneration and death. This study generated multiple clones of induced pluripotent stem cells (iPSCs) and their isogenic controls (ISO) from four patients with MPS II neurodegeneration. MPS II-iPSCs were successfully differentiated into cortical neurons with characteristic biochemical and cellular phenotypes, including axonal beadings positive for phosphorylated tau, and unique electrophysiological abnormalities, which were mostly rescued in ISO-iPSC-derived neurons. RNA sequencing analysis uncovered dysregulation in three major signaling pathways, including Wnt/β-catenin, p38 MAP kinase, and calcium pathways, in mature MPS II neurons. Further mechanistic characterization indicated that the dysregulation in calcium signaling led to an elevated intracellular calcium level, which might be linked to compromised survival of neurons. Based on these dysregulated pathways, several related chemicals and drugs were tested using this mature MPS II neuron-based platform and a small-molecule glycogen synthase kinase-3β inhibitor was found to significantly rescue neuronal survival, neurite morphology, and electrophysiological abnormalities in MPS II neurons. Our results underscore that the MPS II-iPSC-based platform significantly contributes to unraveling the mechanisms underlying the degeneration and death of MPS II neurons and assessing potential drug candidates. Furthermore, the study revealed that targeting the specific dysregulation of signaling pathways downstream of GAG accumulation in MPS II neurons with a well-characterized drug could potentially ameliorate neuronal degeneration.
Similar content being viewed by others
Introduction
Mucopolysaccharidoses (MPS) are inherited lysosomal storage diseases caused by mutations leading to GAG accumulation and cellular dysfunction [1]. Mucopolysaccharidosis type II (MPS II), also known as Hunter syndrome [2], is the most frequent MPS. It is an X-linked recessive disorder predominantly affecting males, resulting from mutations in the IDS gene, which encodes the lysosomal enzyme iduronate-2-sulfatase (IDS). The enzyme hydrolyzes sulfate groups in dermatan sulfate (DS) and heparan sulfate (HS) molecules [3], whose accumulation in MPS II cells disrupts cellular homeostasis and causes organ abnormalities.
The main treatment of MPS II, enzyme replacement therapy (ERT), cannot cross the blood-brain barrier (BBB) [4]. This leaves the severe-form MPS II, which affects the central nervous system, without an effective treatment [5], underscoring the urgency for more dependable models and therapies.
Our study aimed to comprehend the molecular pathogenesis of neurodegeneration and identify potential small-molecule therapeutic candidates. We generated and validated multiple human iPSC clones in four patients with severe-form MPS II, and created mutation-corrected isogenic control (ISO)-iPSCs. These were differentiated into cortical neurons to analyze their biochemical, cellular abnormalities, and electrophysiological malfunctions. Critically, we evaluated the therapeutic effects of various chemicals/drugs in MPS II neurons, targeting dysregulated pathways.
Results
Generation of MPS II-iPSCs and their ISO-iPSC clones
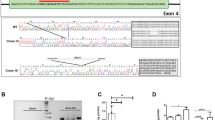
We generated MPS II-iPSCs from four severe-form MPS II patients’ peripheral blood mononuclear cells (PBMCs) using Sendai virus reprogramming. Three optimal clones per patient were selected (Supplementary Fig. 1), exhibiting high pluripotency marker gene expression, silenced Sendai viral genes, normal male karyotypes, and original MPS II mutations (Supplementary Fig. 1A–C). These MPS II-iPSCs also uniformly expressed alkaline phosphatase, TRA-1-60, and NANOG (Supplementary Fig. 1D). Their pluripotency was confirmed through both embryoid body-based differentiation and teratoma formation assays (Supplementary Fig. 1E).
Mutation-corrected ISO-iPSC clones were generated from MPS II-iPSCs through genomic editing, validated by Sanger sequencing (Supplementary Fig. 1F), including two clones each for Patients 1 and 4. NANOG and OCT4 expression confirmed their pluripotency (Supplementary Fig. 1G). The control group comprised these four ISO-iPSC clones and three male healthy control (HC)-iPSC clones. The iPSC clone usage in each experiment was detailed in Supplementary Table 1.
MPS II-iPSCs manifested characteristic MPS II phenotypes
We assessed MPS II phenotypes in MPS II-iPSCs by comparing their IDS activity and GAG content with HC- and ISO-iPSC clones. MPS II-iPSCs showed lower IDS activity (Fig. 1A) and higher GAG content (Fig. 1B), indicating MPS II phenotypes at the pluripotent stem cell stage.
A Deficient IDS activity in MPS II-iPSC clones compared with HC- and ISO-iPSC clones. Each color represents for a unique person. Details of cell clones are provided in the top right corner. B The GAG content in MPS II-iPSC clones was significantly higher than in both control groups. C Representative IF images (upper panels) and flow cytometry results (lower panels) of TUJ1-positive (green) cortical neurons, which were differentiated from the control and MPS II-iPSC clones for four weeks. Nuclei were counterstained by DAPI (blue); Scale bar: 200 μm. D Deficient IDS activity in four-week MPS II neurons compared to HC and ISO counterparts. E The GAG content in four-week MPS II neurons was higher than in their HC and ISO counterparts. F Double IF microscopy showed a significant increase in LAMP-1 signals (red) in four-week MPS II neurons (left). Nuclei were counterstained by DAPI (blue); Scale bar: 20 μm. The histogram (right) shows the quantification results. G Double IF microscopy (left) showed increased Tau-1-positive axonal beading number and area in four-week MPS II TUJ-1-positive neurons compared with control neurons. The arrows indicate beading sites. The histograms show the quantification results of the axonal beading number and area (right) using ImageJ software. DAPI counterstain (blue); Scale bar: 20 μm. For A–E, N = 3 HC clones, N = 4 ISO clones from 4 patients, and N = 12 MPS II clones from 4 patients. For F, G, N = 3 HC clones, N = 4 ISO clones from 4 patients, and N = 9 MPS II clones from 4 patients. All data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.005, between control (either HC or ISO) and MPS II cells, analyzed by Mann–Whitney U test. Each presented data point originates from independent differentiation. Cell clone details are in Supplementary Table 1.
MPS II-iPSC-derived neurons exhibited lysosomal/autophagic abnormalities and axonal beadings
To assess the ability of MPS II-iPSCs to generate neuronal populations, we induced neuronal differentiation in these cells for four weeks. Immunofluorescence (IF) microscopy and flow cytometry demonstrated that most differentiated cells expressed TUJ1 (Fig. 1C), an early neuronal marker [6], and some expressed glial cell marker GFAP (Supplementary Fig. 2A). Neuron and glial proportions were consistent across HC-, ISO-, and MPS II-iPSC clones (Fig. 1C, Supplementary Fig. 2A). Notably, the MPS II-iPSC-derived neurons (hereafter MPS II neurons) exhibited significant IDS deficiency and GAG accumulation compared to HC and ISO neurons (Fig. 1D, E).
During neuronal differentiation, IDS activity increased significantly in HC, ISO, and MPS II neurons (Supplementary Fig. 2B), suggesting its role in this process. Only MPS II neurons accumulated GAGs (Supplementary Fig. 2C). At the fourth week, MPS II neurons exhibited an increase in coarse LAMP-1-positive granules (Fig. 1F). Similarly, western blotting at 15 weeks revealed greater LAMP-2 accumulation and a trend towards increased LAMP-1 in MPS II neurons compared to controls (Supplementary Fig. 2D).
Moreover, the 15-week MPS II neurons exhibited reduced autophagy influx compared to HC and ISO neurons, as evidenced by the reduction in the difference of LC3B levels before and after chloroquine treatment (Supplementary Fig. 2E). Morphologically, increased bead-like swellings were noted in the neurites of 4-week MPS II neurons. IF microscopy confirmed Tau-1 positivity in these axonal beadings (Fig. 1G), with most also co-staining for phosphorylated tau (p-tau) (Supplementary Fig. 2F). Hence, MPS II neurons exhibited lysosomal/autophagic abnormalities of MPS II and an unreported neuropathological trait resembling Alzheimer’s disease (AD) [7].
Anomalies in neurite morphology, cytoskeletal protein expressions, and electrophysiology in long-term cultured MPS II neurons
To characterize neuron morphology, we infected 15-week iPSC-derived neurons with adeno-associated virus (AAV) 8-hSyn-EGFP for visualization (Supplementary Fig. 3A). Fluorescence microscopy revealed abnormal neurite morphology in MPS II neurons compared to HC and ISO neurons (Supplementary Fig. 3A). Image analyses revealed fewer bifurcation nodes, terminal ends, and total branch lengths in MPS II neurons (Supplementary Fig. 3A, Fig. 2A). We examined NEFL, which encodes the neurofilament light chain, expressed earliest [8] and crucial for neurite outgrowth [9]. NEFL mRNA was significantly lower in MPS II neurons, shown by quantitative reverse transcription PCR (qRT-PCR) (Supplementary Fig. 3B). Flow cytometry confirmed reduced NEFL (Fig. 2B), and lower levels of Ankyrin G and spectrin, two proteins essential for neural cytoskeletal integrity and associated with neurodegeneration [10], in MPS II neurons. Whole-cell patch-clamp recording was performed on 18-week iPSC-derived neurons labeled with AAV8-hSyn-EGFP. A decreased percentage of responsive neurons was found in MPS II neurons compared to HC neurons (Fig. 2C, Supplementary Fig. 3C) and ISO neurons (Fig. 2C). In responsive MPS II neurons, increasing current steps (−50–150 pA) caused action potential (AP) anomalies, including prolonged half-width, diminished slopes in 10–90% rise and 90–10% decay, and reduced spike height (Fig. 2C, Supplementary Fig. 3C). To further assay the function of neuronal subtypes, the 20-week iPSC-derived neurons were infected with AAV9-CaMKII α-EYFP and AAV9-mDlx-GFP to label excitatory (the majority) and inhibitory neurons (the minor), respectively (Supplementary Fig. 3D). Both excitatory and inhibitory MPS II neurons exhibited abnormal neurite morphology, with fewer bifurcation nodes compared to HC neurons (Supplementary Fig. 3E, F). Specifically, MPS II excitatory neurons had shorter branch lengths, while inhibitory neurons had fewer terminal ends compared to HC neurons (Supplementary Fig. 3E, F). Electrophysiologically, in 22-week MPS II excitatory neurons, there was a slightly reduced percentage of neurons responsive to current injection compared to HC neurons (Supplementary Fig. 3G). They also displayed abnormal AP, with decreased spike height, prolonged half-width, and reduced 10–90% rising slope compared to HC neurons (Supplementary Fig. 3G).
A Neurite morphometrics of mature cortical neurons. The 15-week MPS II, HC, and ISO neurons were infected with AAV8-hSyn-EGFP. Neuron morphology was reconstructed using Neurolucida. The analyzed neuron morphology parameters included soma size, segments, bifurcation nodes, terminal ends, and total branch lengths. N = 3 HC clones, N = 4 ISO clones from 2 patients, and N = 12 MPS II clones from 4 patients. B Flow cytometry analyses demonstrated decreased NEFL, Ankyrin G, and Spectrin protein levels in 15-week MPS II neurons. N = 6 HC clones, N = 6 differentiations from 2 patients’ 4 ISO clones, and N = 6 MPS II clones from 4 patients. C Abnormal action potentials (APs) in mature MPS II neurons compared to HC and ISO neurons. The APs of cortical neurons (differentiated for 18 weeks from control and MPS II iPSCs) in response to increasing current steps (−50 to 150 pA) were measured using current-clamp recordings. A decreased response rate to current injections in MPS II neurons compared to those of HC and ISO neurons (upper left panel). N = 14 cells of 4 differentiations of HC-1, N = 15 cells from 4 differentiations of ISO-4, and N = 24 cells from 3 differentiations of MPS II 4-1. ***p < 0.005, analyzed by chi-square test. Representative traces depicted the changes of AP morphology in MPS II neurons compared with HC and ISO neurons. Scale bars: 10 ms (x-axis) and 10 mV (y-axis). Histograms show the differences of AP parameters between MPS II neurons and HC or ISO neurons, including AP threshold, AP spike height, AP half-width, after-hyperpolarization (AHP), the slope of 10–90% rise, and the slope of 90–10% decay. Each data point represents an individual responsive neuron. N = 12 cells from 4 differentiations of HC-1, N = 13 cells from 4 differentiations of ISO-4, and N = 6 cells from 3 differentiations of MPS II 4-1. Data represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.005, analyzed by Mann–Whitney U test. Cell clone details are in Supplementary Table 1.
Identification of dysregulated genes and pathways in MPS II neurons
To explore MPS II neurodegeneration and identify specific gene expression changes, we conducted RNA sequencing on 15-week HC neurons (3 clones) and MPS II neurons (9 clones), identifying 921 upregulated and 649 downregulated differentially expressed genes (DEGs) in MPS II neurons.
Ingenuity Pathway Analysis (IPA) was employed to investigate the DEGs that might influence the neuronal AP, identifying three ion channel-encoding genes: SCN9A (sodium channel NaV1.7) [11], KCNMB2 [12], and KCND2 (potassium channels) [13]. The qRT-PCR confirmed their lower expression in MPS II neurons (Supplementary Fig. 3H). Flow cytometry showed lower protein levels of these genes in MPS II neurons (Supplementary Fig. 3I), which might contribute to the AP anomalies. Furthermore, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis revealed significant DEG enrichment in several pathways, with Wnt ranking highest, followed by calcium and MAPK signaling pathways in MPS II neurons (Fig. 3A).
A KEGG pathway analysis of DEGs in MPS II neurons compared to HC neurons revealed significant changes in multiple signaling pathways. B GSEA of DEGs in MPS II neurons revealed significant enrichment of a gene set involved in negative regulation of the canonical Wnt signaling pathway (left), and a gene set that was negatively correlated with Wnt upregulation (right) in 15-week-old MPS II neurons compared with their HC counterparts. C Decreased GSK-3β phosphorylation and increased β-catenin phosphorylation in 15-week-old MPS II neurons compared with control neurons. The upper panel shows a representative western blotting result, comparing the phosphorylation levels of GSK-3β at Ser9 and β-catenin at Ser33/37/Thr41 among HC, ISO, and MPS II neurons. The lower panels depict the relative phosphorylation levels of GSK-3β (left) and β-catenin (right) in different groups of neurons. The levels resulted from normalizing the phosphorylation signals to respective total GSK-3β or β-catenin signals, as well as GAPDH signals, and are presented as mean ratios relative to HC neurons. N = 8 differentiations from 2 HC clones, N = 8 differentiations from 4 ISO clones, and N = 12 differentiations from 6 MPS II patients’ clones. D The qRT-PCR analysis confirmed the altered expression of five representative Wnt pathway genes, including five well-known upstream genes (NOTUM, WNT7B, RSPO3, FZD9, and GPC3) and four downstream genes (ABI1, UPP1, MMP7, and FOSL1) in 15-week-old MPS II neurons compared to their control counterparts. N = 3 HC clones, N = 4 ISO clones, and N = 7 differentiations from 4 MPS II patients’ clones. E Representative dot plots for the flow cytometry analysis of phosphorylated p38 MAPK-positive neurons. Histograms for the flow cytometry show a decreased percentage of phosphorylated p38 MAPK (p-p38)-positive cells, and a lower mean fluorescence intensity (MFI) of p-p38 MAPK GFP signals in 15-week MPS II cortical neurons compared with their control counterparts. N = 3 HC clones, N = 5 differentiations from 4 ISO clones, and N = 7 differentiations from 4 MPS II patients’ clones. *p < 0.05, **p < 0.01, analyzed using chi-square test for positive cell percentage data, and Mann–Whitney U test for MFI data. F Representative IF microscopy images of p-p38 MAPK-positive iPSC-derived neurons. Quantification showed a significantly decreased nuclear-to-cytoplasmic ratio of p-p38 MAPK fluorescence intensity (left histogram), and a lower percentage of nuclei positive for p-p38 MAPK (right histogram) in 15-week MPS II neurons compared to their control counterparts. DAPI counterstain: blue. Scale bar: 20 μm. The intensity of nuclear and cytoplasmic p-p38 signals and the percentage of p-p38 MAPK-positive nuclei were quantified using ImageJ. N = 3 HC clones, N = 4 ISO clones, and N = 3 MPS II patients’ clones. **p < 0.01, ***p < 0.005, analyzed using Welch’s test for fluorescence intensity ratio data, and chi-square test for nuclei positivity percentage data. Data in other panels represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.005, analyzed by Mann–Whitney U test. Cell clone details are in Supplementary Table 1.
Additionally, gene set enrichment analysis (GSEA) demonstrated genes in MPS II neurons significantly enriched in negative regulation of Wnt pathway (Fig. 3B, left) and downregulation of WNT3A-responsive genes (Fig. 3B, right). KEGG pathway analysis revealed that upregulated genes in Wnt signaling were mostly upstream regulators, while downregulated genes were downstream TCF/LEF targets (Supplementary Fig. 4A). These suggested attenuated intracellular Wnt signaling in MPS II neurons, with upstream regulator upregulation possibly compensating for this attenuation. Supporting this, western blotting demonstrated decreased phosphorylated GSK-3β (Ser9, p-GSK-3β, inactive), and increased β-catenin phosphorylation (Ser33/37/Thr41, p-β-catenin, inactive) in MPS II neurons (Fig. 3C). It also revealed less nuclear and more cytosolic non-phosphorylated β-catenin (active) in MPS II neurons (Supplementary Fig. 4B). Next, we analyzed nine established Wnt signaling genes from the DEGs (Supplementary Fig. 4C) and literature [14,15,16] using qRT-PCR, confirming upregulation of five upstream genes (NOTUM, WNT7B, RSPO3, FZD9, and GPC3 [14]) and downregulation of four downstream genes (ABI1 [15], UPP1 [16], FOSL1, and MMP7) in MPS II neurons (Fig. 3D). Additionally, the 15-week iPSC-derived neurons transduced with Wnt reporter construct-containing lentiviruses showed significantly lower Wnt reporter activity in MPS II neurons compared to control neurons (Supplementary Fig. 4D). These findings collectively confirmed reduced Wnt/β-catenin signaling in MPS II neurons.
IPA showed that p38 MAPK signaling was significantly altered in MPS II neurons (Supplementary Table 2), consistent with dysregulated MAPK pathway in KEGG pathway enrichment analysis (Fig. 3A) and the suppressed fibroblast growth factor (FGF) signaling disclosed by GSEA (Supplementary Fig. 4E). Concordantly, flow cytometry (Fig. 3E) showed reduced phosphorylated p38 (p-p38) MAPK in 15-week MPS II neurons, both in cell percentage and fluorescent intensity. IF microscopy showed a lower nuclear-to-cytoplasmic p-p38 ratio and fewer p-p38 MAPK-positive nuclei in MPS II neurons (Fig. 3F). This trend was also observed in total p38 MAPK (Supplementary Fig. 4F). Furthermore, MPS II neurons responded less to Wnt activator WNT3A and p38 MAPK activator bFGF2, showing decreased activation of downstream genes (AXIN2, MYC, and MMP7 for WNT3A [17]; FOSL1 [18], CCDC115 [19], and OTX2 [20] for FGF2) (Supplementary Fig. 4G, H).
Dysregulated calcium homeostasis in MPS II neurons and the possible mechanisms
KEGG pathway analysis identified calcium signaling as the second most influenced pathway in MPS II neurons (Fig. 3A). GSEA further confirmed this, highlighting enriched genes associated with cytosolic calcium ion concentration, positive regulation of calcium ion sequestration, and calcium channel complex in MPS II neurons (Fig. 4A, Supplementary Fig. 5A). KEGG pathway mapping (Fig. 4B) showed these enriched genes as upregulated or downregulated across calcium pathway nodes, most verified by qRT-PCR (Fig. 4C). Fura-2 calcium imaging and spectrofluorometry both indicated higher baseline intracellular calcium levels in MPS II neurons, even pre-treatment (Fig. 4D, Supplementary Fig. 5B). After Thapsigargin (TG) treatment, MPS II neurons exhibited greater calcium release than controls, suggesting disturbed endoplasmic reticulum (ER) calcium homeostasis (Fig. 4E, Supplementary Fig. 5B).
A GSEA of RNA-seq data indicated significant enrichment of the genes regulating cytosolic calcium ion concentrations (left panel) and positive regulation of calcium ion sequestering (right panel) in MPS II neurons (N = 9) compared with HC neurons (N = 3). B Comparative analysis of RNA-seq data between the 15-week MPS II and HC neurons highlighted a change in the calcium signaling pathway (hsa04020 in KEGG) in MPS II neurons. The mapping of the regulated genes in the pathway suggested a potential alteration in the calcium homeostasis in MPS II neurons. N = 3 clones for HC, N = 9 clones from 4 patients for MPS II. C The RNA-seq data were validated by qRT-PCR, confirming the majority of expression perturbations in several calcium signaling-related genes, as shown in A, in MPS II (N = 5 differentiations from 4 patients’ clones) neurons compared with HC (N = 4 differentiations from 3 clones) and ISO (N = 4 differentiations from 2 ISO clones from 2 patients) neurons. D MPS II neurons showed elevated baseline calcium levels compared to HC neurons. The intracellular calcium changes were quantified by assessing the fluorescence intensity of the Fura-2 dye at 340 nm and 380 nm wavelengths using fluorescence microscopy in 15-week HC (N = 4 differentiations from 2 clones), ISO (N = 4 differentiations from 2 ISO clones from 2 patients), and MPS II (N = 8 differentiations from 4 clones from 3 patients) neurons. Each dot in the histogram represents the mean of 13 time points for a single differentiated clone. E Thapsigargin (TG) treatment caused a greater increase in intracellular calcium levels in MPS II neurons. The differences in the fluorescence intensity ratios at 340 nm and 380 nm (340/380) after TG treatment were determined by subtracting the baseline 340/380 ratio from the post-treatment values. This difference is represented as “∆ (340/380)”. The arrow indicates the moment when TG was added. The area under the curve (AUC) of the ∆ (340/380) over a 10-min period was computed, providing a quantitative measure of changes in intracellular calcium concentration (right panel). F A calcium channel blocker, verapamil, rescued the altered expression of certain genes in B that regulate calcium signaling (CACNA1A, P2RX3, ADRA1A, and PLCD3), and the genes encoding other ion channels (SCN9A and KCND2) and a neurofilament protein (NEFL) in 15-week MPS II neurons (N = 4 differentiations from 4 patients’ clones). The data represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.005, analyzed by Mann–Whitney U test for A, C–E, and Wilcoxon signed-rank test for F. Cell clone details are in Supplementary Table 1.
To further explore dysregulated calcium signaling, we treated 15-week HC neurons with excessive HS, a GAG accumulated in MPS II, revealing upregulation of calcium channel genes CACNA1A and P2RX3 (Supplementary Fig. 5C). Conversely, GSK-3β inhibitors, CHIR99021 or Tideglusib, downregulated these genes in MPS II neurons (Supplementary Fig. 5D). Moreover, Ionomycin, a calcium ionophore, caused gene expression changes in HC neurons resembling MPS II neurons, including downregulation of ADRA1A, PLCD3, MYLK2, PDE1B, SCN9A, and KCND2 (Supplementary Fig. 5E). In contrast, verapamil, a calcium channel blocker, reversed many gene expression alterations in MPS II neurons (Fig. 4F). Therefore, both HS accumulation and GSK-3β overactivity might contribute to the elevated intracellular calcium and altered calcium pathway gene expression observed in MPS II neurons. This could be rescued by a readily accessible calcium channel blocker drug.
Selected drug candidates rescued abnormalities in MPS II neurons
MPS II neurons were employed to test various potential therapeutic agents: rhIDS (MPS II drug); A-443654 (predicted by the LINCS L1000 data repository [21] to rescue the expression of GPC3, MMP7, and NEFL); Baicalein (Tau-1 aggregation inhibitor [22]); Tideglusib (GSK-3β inhibitor [23]); Odiparcil (GAG-clearing drug [24]); surfen (HS antagonist [25]); and NK-1 (HS/DS binder [26]). Except for rhIDS and Odiparcil, all can cross the BBB [22, 23, 25,26,27]. Drug dosages and durations were determined based on literature [23,24,25,26,27,28] and our cytotoxicity test (Supplementary Table 3), applied to four-week and 15-week MPS II and control neurons (Supplementary Fig. 6A). Among them, rhIDS, baicalein, and Tideglusib, significantly reduced GAGs (Fig. 5A). Only rhIDS enhanced IDS activity when administered alone (Supplementary Fig. 6B, C), with Tideglusib further boosting it in four-week (Fig. 5B), but not in 15-week neurons (Supplementary Fig. 6D). Nevertheless, combined rhIDS and Tideglusib treatment from weeks four to six, followed by rhIDS alone until week 15, improved IDS activity in MPS II neurons more than rhIDS alone (Supplementary Fig. 6E). Significant viability improvement was observed in the 15-week MPS II neurons treated with A-443654, baicalein, Odiparcil, or Tideglusib for 24 h (Fig. 5C), while rhIDS required a longer incubation (seven days) for viability enhancement (Supplementary Fig. 6F). Furthermore, qRT-PCR showed that rhIDS or Tideglusib significantly upregulated NEFL and MMP7, and downregulated GPC3 in 15-week MPS II neurons (Fig. 5D–F). Additionally, Tideglusib decreased Tau-1 and phosphorylated tau in axon-beading regions of 15-week MPS II neurons (Fig. 5G, H). These results suggested that Tideglusib, with its multifaceted rescuing effects, was the most promising agent tested.
MPS II neurons were used to test the therapeutic effects of human rhIDS (20 ng/mL) and several potential drugs, including A-443654 (0.05 μM), baicalein (10 μM), Odiparcil (10 μM), surfen (10 μM), NK-1 (0.5 μM), and Tideglusib (10 μM). Solvents (vehicles) alone were used as the control. A GAG colorimetric assay was performed to measure the GAG content in cells that were treated with drugs for seven days. B IDS activity assay was performed in four-week MPS II neurons treated with rhIDS (20 ng/mL) alone or in combination with different drugs for seven days to evaluate the effects of the drugs on IDS activity. C The effect of drugs on the viability of MPS II neurons was assessed by flow cytometry using the live cell indicator calcein-AM after 24 h of drug treatment. The qRT-PCR showed the differential rescue effect of drugs on the mRNA levels of marker genes, such as the downregulated NEFL in D, or the DEGs of RNA sequencing, including MMP7 in E and GPC3 in F in the 15-week MPS II neurons treated with different drugs for 24 h. The mRNA levels were normalized to GAPDH and presented as fold change relative to vehicle-treated cells. All data were compared with the control vehicle, except in B, where they were compared with IDS alone, and both analyzed by Student’s t test. G, H IF microscopy showed that 15-week MPS II Tuj-1-positive neurons exhibited significantly increased numbers and areas of Tau-1- (G) and p-tau- (H) positive axonal beadings. Furthermore, the Tideglusib (10 μM) treatment rescued the phenotypes. Scale bar, 20 μm. Insets: magnified images of the beading regions (dotted squares). N = 3 clones from 3 patients for MPS II in A–F; N = 3 independent differentiations from 1 clone for HC and ISO, and N = 12 independent differentiations from 2 patients’ clones for MPS II in G and H. Data represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.005, analyzed by paired Student’s t test for A, B, D–F, chi-square for C, and Mann–Whitney U test for G, H. Each data point presented originates from independent differentiations. Descriptions of cell clones used are provided in Supplementary Table 1.
Tideglusib reversed functional and morphological aberrations in MPS II neurons
In 15-week MPS II neurons, Tideglusib effectively corrected the aberrant expressions of the majority of aforementioned calcium pathway genes and the three AP-related DEGs: SCN9A, KCNMB2, and KCND2 (Fig. 6A, Supplementary Fig. 5D).
A Tideglusib treatment (10 μM, 1 day) reversed the abnormal downregulation of the genes associated with calcium signaling pathways (PLCD3, MYLK2, and PDE1B) and genes encoding sodium or potassium channels (SCN9A, KCNMB, and KCND2), and reversed the abnormal upregulation of the phospholipase-encoding PLCB2 gene in 15-week MPS II neurons (N = 6 clones from 4 patients). B Tideglusib mitigated the intracellular calcium increase caused by TG treatment in MPS II neurons. The intracellular calcium changes were quantified by assessing the fluorescence intensity of the Fura-2 dye at 340 nm and 380 nm wavelengths using fluorescence microscopy in 15-week MPS II neurons and categorized as vehicle control (N = 9 differentiations from 6 clones), Tideglusib pretreatment (N = 9 differentiations from 6 clones), and Tideglusib intra-treatment (N = 9 differentiations from 6 clones). To assess the effect of treatment, we subtracted the baseline 340/380 ratios from the post-treatment fluorescence intensity ratios, calculating the change, namely Δ (340/380). The Tideglusib pretreatment and Tideglusib intra-treatment groups both showed reduced ∆ (340/380) values compared to the Vehicle group. The area under the curve (AUC) of the ∆ (340/380) over a span of 10 min was computed, providing a quantitative depiction of intracellular calcium concentration changes. The arrow indicates the time when TG alone or TG combined with Tideglusib was added. Vehicle pretreatment: MPS II neurons pre-treated with the vehicle prior to TG treatment; Tideglusib pretreatment: MPS II neurons pre-treated with Tideglusib for 24 h prior to TG addition; Vehicle intra-treatment: MPS II neurons treated with TG and vehicle; Tideglusib intra-treatment: MPS II neurons treated simultaneously with both Tideglusib and TG. C The rescuing effect of Tideglusib on the electrophysiology of MPS II neurons. The 15-week MPS II 1-2 neurons were treated with 10 μM Tideglusib or vehicle (DMSO) for 2 weeks. The APs in MPS II neurons, as mentioned above, were measured using current-clamp recordings in response to increasing current steps (−50 to 150 pA). Tideglusib treatment trended toward an increased recordable AP percentage in MPS II neurons (left upper panel). N = 16 cells from 3 independent differentiations of 1 clone for vehicle group; N = 14 cells from 5 independent differentiations of 1 clone for Tideglusib group. p = 0.2, analyzed by chi-square test. Representative change of the AP morphology of MPS II neurons treated with Tideglusib compared to those treated with vehicle (right upper panel). Scale bars: 10 ms (x-axis) and 10 mV (y-axis). Histograms show the differences of AP parameters between vehicle- and Tideglusib-treated MPS II neurons, including AP threshold, AP spike height, AP half-width, after-hyperpolarization (AHP), the slope of 10–90% rise, and the slope of 90–10% decay. N = 12 cells from 3 independent differentiations for vehicle group; N = 12 cells from 5 independent differentiations for Tideglusib group. D The rescuing effect of Tideglusib on the neurite morphology of MPS II neurons. The 15-week MPS II neurons were treated with 10 μM Tideglusib or vehicle (DMSO) for 2 weeks. Neurite morphometrics were measured using Neurolucida Neuron Tracing software. Representative images of the 15-week MPS II neurons transduced with AAV9-CaMKII α-EYFP. Neuron morphology through fluorescence microscopy is displayed in the upper panels, while their reconstructed images by Neurolucida are shown in the lower panels. Scale bar, 10 μm. The morphometric parameters of neurites included soma size, segments, bifurcation nodes, terminal ends, and total branch lengths. N = 4 clones from 2 MPS II patients for each group. Data represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.005, analyzed by Wilcoxon signed-rank test for A, B, and D, Mann–Whitney U test for C. Cell clone details are in Supplementary Table 1.
Concordantly, Fura-2 calcium imaging demonstrated that Tideglusib significantly reduced abnormal calcium-releasing responses to TG in MPS II neurons, both in pre- and simultaneous treatments (Fig. 6B). To assess Tideglusib’s therapeutic effects on electrophysiology and neurite morphology, we established an optimal regimen for 15-week neurons (Supplementary Fig. 6G), confirming 10 µM Tideglusib’s efficacy over two weeks by activating the canonical Wnt target gene AXIN2 [29] (Supplementary Fig. 6H), without affecting neuronal survival. Patch-clamp recordings indicated a trend of increased responsiveness in MPS II neurons and significant improvements in AP threshold, AP half-width, and 90–10% decay slope post-Tideglusib (Fig. 6C). Morphometric analysis showed that Tideglusib increased bifurcation nodes, terminal ends, and total branch lengths in MPS II neurons (Fig. 6D).
Regulation of ion channel genes in iPSC-derived neurons by p38 MAPK and Wnt/β-catenin signaling
Finally, we investigated if decreased expression of ion channel genes (KCND2, KCNMB2, and SCN9A) in MPS II neurons was directly associated with dysregulated p38 MAPK or Wnt/β-catenin signaling. Treating iPSC-derived neurons with SB203580 (p38 MAPK inhibitor) or MSAB (Wnt/β-catenin inhibitor) downregulated these genes and a known common target gene (FOSL1) across all neuron groups (Supplementary Fig. 7A). Conversely, Anisomycin (p38 MAPK activator) or Tideglusib (Wnt/β-catenin activator) upregulated these genes, particularly SCN9A (Supplementary Fig. 7A). Concordantly, chromatin immunoprecipitation (ChIP) analysis confirmed β-catenin binding to the SCN9A promoter LEF1/TCF consensus sequence in 15-week neurons (Supplementary Fig. 7B). This suggested that dysregulated Wnt/β-catenin signaling in MPS II neurons might directly downregulate key ion channel genes, contributing to abnormal neuronal APs.
Discussion
In the past decade, ERT has notably improved life expectancy and quality for MPS II patients, yet it falls short in addressing severe-form MPS II cases, where neuropsychiatric abnormalities persist despite ERT [5]. New approaches like intrathecal rhIDS face risks like device malfunction [4]. Izcargo® (pabinafusp alfa) exploits transcytosis for rhIDS to cross the BBB, showing initial neurocognitive improvement [30], but long-term effects are unclear. Meanwhile, alternative treatments like hematopoietic stem cell transplantation [31] and gene therapy [30], encounter efficacy debates [5] or clinical trial challenges, including complications, immunogenicity, and costs [30].
MPS II-iPSC clones have been reported in literature [32,33,34,35,36], and some were differentiated into neuronal populations, displaying phenotypes like GAG accumulation, increased LAMP-2 expression, apoptosis, ER and Golgi complex structural changes, and secondary lipid accumulation. However, our models, employing more mature neurons, revealed unique and previously unreported findings in MPS II neurons, including morphometric and electrophysiological abnormalities, transcriptome variations, signaling pathway dysregulation, and potential therapeutic agents. An important abnormality in MPS II neurons was axonal beading, reminiscent of axonal spheroids seen in other neurodegenerative conditions [37]. Impairment of axonal transport has been suggested as a likely cause of axonal spheroids [38]; however, further investigation is needed to ascertain its relevance in our system. Our results indicated that these abnormalities were Tau-1 positive varicosities, linked to tau aggregation. This new phenotype is reasonable since tau aggregation depends on HS [39], a GAG accumulated in MPS II. Notably, over-sulfated HS in MPS II has been hypothesized to enhance tau aggregation [40], as sulfated HS is necessary for tau’s binding to HS proteoglycans [41]. Accumulation of hyperphosphorylated tau has been reported in the brains of MPS IIIA patients [42] and MPS IIIB mice [43]. Whether tau aggregations exist in human MPS II brain pathological specimens remains to be determined, but this raises the possibility of using currently available tau-aggregation inhibitors to treat MPS II neurodegeneration, provided tau aggregations are proven to exist. Congruently, our results showed that baicalein, a tau-aggregation inhibitor, partially reversed MPS II neuron phenotypes. Notably, Tideglusib treatment also reduced axonal varicosities, indicating GSK-3β inhibition may lessen tau aggregation in MPS II, similar to its effect in AD [44]. However, in AD brains, elevated GSK-3β stems from Aβ [45], while in MPS II neurons, reduced Wnt and p38 MAPK signaling might be the cause.
The p38 MAPK signaling interacts with the Wnt/β-catenin signaling in various ways. For instance, WNT3A activates p38 MAPK through Dishevelled [46], while p38 MAPK phosphorylates GSK-3β‘s C-terminus, increasing β-catenin nuclear accumulation [47]. Additionally, p38 MAPK phosphorylates myocyte enhancer factor 2 (MEF2), enhancing MEF2’s nuclear translocation and interaction with β-catenin, activating Wnt and MEF2 target genes [48]. Moreover, AP-1, another p38 MAPK effector, synergistically activates Wnt/β-catenin target genes with β-catenin/TCF [49]. Wnt and p38 MAPK cooperate in negatively regulating autophagy. Both inhibit GSK-3β, and p38 MAPK hampers autophagy by phosphorylating ULK1 [50], a GSK-3β-activated kinase [51] that triggers autophagosome formation. Conversely, autophagy downregulates Wnt signaling by degrading Dishevelled [52] and suppresses p62-mediated p38 MAPK activation [53].
GAG accumulation in our MPS II cells was milder at the iPSC stage but worsened during neuronal differentiation, which could explain the non-significant increase in GAG levels in MPS II iPSCs compared to ISO iPSCs. Similarly, Rybová et al. [33] also found no significant increase in GAG levels in MPS II iPSCs compared to healthy controls, suggesting a physiological role of enhanced GAG synthesis during neuronal differentiation. Undegraded GAG accumulation can affect cell signaling [54]. Costa et al. reported increased Wnt signaling in Ids morpholino-treated zebrafish heart valves [55], and Salvalaio et al. found altered Wnt signaling in Ids-knockout mouse brains, with marked regional variations [56]. Our results concurred with the mixed Wnt gene regulation (both upregulation and downregulation). However, the expression of Wnt downstream genes was downregulated in our results, but unchanged in theirs [56], possibly due to differences in neuronal maturation stages and species. Reduced Wnt/β-catenin signaling led to some downstream genes’ reduced expression, which might have prompted the upregulation of specific upstream genes, compensating for the weakened signaling.
Although the precise mechanisms of MPS II neurodegeneration remain unclear, our study proposes an intriguing model (Fig. 7) for the disease’s pathogenesis. The scenario might begin with intracellular accumulation of GAGs in lysosomes, causing lysosomal destabilization, initial autophagy induction, and subsequently autophagic buildups with increased LAMP-2 expression. Initial autophagy induction could weaken both Wnt [52, 57] and p38 MAPK [53] signaling. Extracellularly, GAGs, especially HS affected growth factor distribution and function due to their high affinity with these molecules [58]. Excessive HS-proteoglycans in MPS I and IIIB cells have been shown to sequester FGF2, reducing its signaling, which is reversible with HS-targeting proteins [59]. Jenkins et al. [60] found that Wnts’ interaction with HS in membrane proteoglycans inhibits Wnt signaling by trapping Wnt ligands. Excessive GAGs may thus sequester Wnt ligands and growth factors, decreasing their binding to frizzled receptors and receptor tyrosine kinases (RTK), reducing both Wnt and RTK-p38 MAPK signaling (Fig. 7A). This led to increased GSK-3β activity due to reduced inhibition, further suppressing Wnt/β-catenin signaling. Decreased p38 MAPK activity downregulated its downstream genes, including those shared by Wnt/β-catenin signaling (FOSL1) and ion channel genes (SCN9A, KCNMB2, and KCND2), crucial for APs (Fig. 7B) [61, 62]. Meanwhile, elevated intracellular calcium is also known to raise the AP threshold and suppress neuron excitability by activating several calcium-gated potassium currents [63]. Additionally, it also contributes to neuronal death [64].
A Defective degradation of GAGs in lysosomes leads to a decline in autophagy flux and a consequent increased accumulation of LAMP-2. Furthermore, excessive extracellular storage of GAGs results in an increased accumulation of HSPGs on the cell membrane. The excessive GAG chains on HSPGs in MPS II neurons may sequester specific ligands (such as Wnts and growth factors), thereby depressing the activation of Wnt and RTK receptors. The decrease in Wnt receptor-mediated signaling enhances GSK-3β activity. This, in turn, suppresses autophagy flux, promotes tau aggregation, and elevates intracellular calcium concentration via activation of the expression of selective genes that encode calcium channels. Additionally, it inhibits β-catenin/TCF-regulated gene expression (e.g., AXIN2, MMP7, and NEFL). On the other hand, reduced RTK signaling diminishes the active (phosphorylated) form of p38 MAPK and its nuclear translocation, thereby reducing both p38 MAPK-mediated inhibitory phosphorylation on GSK-3β and p38 MAPK-activated gene expression (such as FOSL1 and genes encoding selected ion channels, e.g., SCN9A). The width of the arrows indicates the relative strength of regulation, and the dashed lines indicate decreased levels of nuclear translocation. Genes regulated by both p38 MAPK and β-catenin/TCF are denoted in blue font. B Decreased p38 MAPK and Wnt/beta-catenin signaling in MPS II neurons may lead to abnormal AP via ion channel gene dysregulation (e.g., SCN9A, KCNMB2, and KCND2). Tideglusib treatment restores the expression of these genes and mitigates the AP anomalies of MPS II neurons. Red arrows indicate increased expression; blue arrows indicate decreased expression.
Dysregulated calcium homeostasis in MPS II neurons in calcium imaging aligned with calcium pathway gene alterations in KEGG pathway mapping. Upregulated genes like CACNA1A, P2RX3, and PLCB2 might increase baseline cytosolic calcium and ER calcium release upon TG treatment in calcium imaging. Lysosomal storage diseases, like Gaucher disease [65] and Niemann-Pick type C1 [66], demonstrate how accumulated substrates can disrupt calcium homeostasis by affecting ryanodine receptors and calcium ATPases/Na+/Ca2+ exchangers, respectively.
Likewise, accumulated GAGs in MPS II neurons might influence calcium channels or calcium-handling proteins. This was supported by our qRT-PCR results, showing upregulation of CACNA1A and P2RX3 after excessive HS treatment, linking HS accumulation to calcium homeostasis changes. Moreover, increased GSK-3β activity might also contribute. GSK-3β is known to interact with inositol trisphosphate receptor, influencing its phosphorylation and calcium release from ER [67]. GSK-3β overexpression in mice correlates with reduced Serca2a expression and elevated cytosolic calcium in cardiomyocytes [68]. Thus, elevated GSK-3β in MPS II neurons likely altered calcium homeostasis. Our qRT-PCR supported this, showing that GSK-3β inhibitors downregulated CACNA1A and P2RX3.
Other gene expression changes in the KEGG calcium pathway might be secondary or compensatory. Elevated intracellular calcium may upregulate MYLK2 [69] and PDE1B [70] enzyme activities, causing their mRNA compensatory downregulation. Supporting this, Ionomycin-induced calcium elevation downregulated MYLK2 and PDE1B in control neurons. Additionally, PLCD3 downregulation is a secondary effect of increased intracellular calcium [71], as supported by Ionomycin’s effect on this gene.
Our results revealed Tideglusib’s effectiveness in correcting morphological and electrophysiological aberrations of mature MPS II neurons. Tideglusib, a selective non-ATP-competitive GSK-3β inhibitor [72], has been applied in clinical trials for several neurological diseases, including AD [73], myotonic dystrophy type 1 [74], progressive supranuclear palsy [75], and amyotrophic lateral sclerosis [76]. This suggests the involvement of GSK-3β in a fundamental pathway shared among multiple neurodegenerative disorders. However, its definite efficacy and optimal dosing require further validation in larger clinical trials. Given its ability to penetrate the BBB [23] and its established human pharmacokinetics and safety profile [77], Tideglusib holds significant potential for treating MPS II neurodegeneration, either alone or in combination with other therapies, pending validation through animal studies.
This study unveils novel morphological and functional abnormalities in mature MPS II neurons, and their mechanistic association with dysregulated signaling pathways. It highlights Tideglusib, a GSK-3β inhibitor, as a promising candidate for correcting neuronal phenotypes, offering valuable insights for MPS II therapy. These discoveries advance our comprehension of MPS II neurodegeneration and suggest new therapeutic strategies.
Materials and methods
Human subjects and sample collection
Blood was collected from four male patients with the severe form of MPS II diagnosed at the MacKay Memorial Hospital in Taiwan based on their clinical features, low IDS enzyme activities, and pathogenic mutations of the IDS gene, with their parents’ written informed consent. The protocol was approved by the Research Ethics Committee of both the National Taiwan University Hospital and MacKay Memorial Hospital. The ages and IDS mutations of these patients were as follows: Patient 1, 29 years, CGG > TGG (R468W) point mutation; Patient 2, 14 years, 1214_1215 del CT; Patient 3, 20 years, IVS7+5G>C; Patient 4, 28 years, c.1122C>T, (p.G374G). The collected blood in heparin-containing tubes was mixed with Histopaque (Sigma-Aldrich, MA, USA) at a 1:1 ratio, and the PBMCs were isolated by centrifugation at 400 × g for 30 min at room temperature.
Generation of MPS II-iPSC
The iPSCs were derived using a nonintegrated Sendai virus system (CytoTune™-iPS 2.0 Sendai Reprogramming Kit, Thermo Fisher Scientific, MA, USA) according to the manufacturer’s instructions. Briefly, MPS II-PBMCs were plated at a density of 1 × 106 cells/well in four well plates in StemPro®-34 SFM medium (Thermo Fisher Scientific, MA, USA) supplemented with cytokines (100 ng/mL stem cell factor, 100 ng/mL FMS-like tyrosine kinase 3, 20 ng/mL IL-3, 20 ng/mL IL-6) and Glutamax (Thermo Fisher Scientific, MA, USA). Half of the culture medium was replaced each day for four days, and a combination of Sendai virus expressing human OCT3/4, SOX2, KLF4, and MYC was added to the target cells. Three days later, the infected PBMCs were replated onto mitomycin-C-inactivated MEFs. Two days later, the StemPro®-34 SFM medium (Thermo Fisher Scientific, MA, USA) without cytokines was changed for two days and then switched to StemFlex (Thermo Fisher Scientific, MA, USA) medium. The iPSC clones grown for three weeks were then selected, expanded, and subjected to further characterization, as described below. Finally, three optimal iPSC clones that fulfilled the criteria for human iPSCs [78] were selected for each patient and labeled as -1, -2, and -3.
Healthy male control iPSCs and the culture of iPSCs
Healthy male control iPSC clones (HC-1~3), which were obtained from the Bioresources Collection and Research Center (BCRC), Taiwan, with the original names of IBMS-iPSC-02-07 (SC81031), TVGH-iPSC-02-07 (SC81033), and NTUH-iPSC-02-02 (SC81035), respectively, were reprogrammed by Sendai virus as human iPSC clones from three male subjects, generated by the Human Disease Induced Pluripotent Stem Cell Service Consortium (National Core Facility for Biopharmaceuticals, Taiwan). All iPSCs were cultured on Matrigel (Corning, NY, USA) with StemFlex (Thermo Fisher Scientific, MA, USA) at 37 °C in a humidified atmosphere containing 5% CO2.
In vitro differentiation of MPS II-iPSCs
For in vitro differentiation, MPS II-iPSC colonies were dispersed into small clumps using dispase (Thermo Fisher Scientific, MA, USA; 2 mg/ml for 10 min) and transferred onto ultralow attachment plates (Corning, NY, USA) for embryoid body (EB) formation in iPSC medium, which was changed every day for four days. The EBs were then transferred to culture dishes coated with 0.1% gelatin and fetal bovine serum (FBS)-containing medium. During the 14-day differentiation period, the medium was changed every two days.
Teratoma formation assay
Approximately 1–1.5 × 107 MPS II-iPSC cells were injected subcutaneously into the backs of 6–8-week-old BALB/c nude mice (National Laboratory Animal Center, Taipei, Taiwan). Teratomas were allowed to develop for up to 8–10 weeks and were then excised and fixed with 4% paraformaldehyde overnight at 4 °C before embedding in paraffin for sectioning. The samples were cut to 7 µm in thickness and stained with hematoxylin and eosin stain (H&E) for histological analysis. All animal experiments were undertaken according to the Guidebook for the Care and Use of Laboratory Animals (published by the Chinese-Taipei Society of Laboratory Animal Sciences) and approved by the Institutional Animal Care and Use Committee at National Taiwan University, College of Medicine. The well-being of the animals was monitored daily, and the animals had free access to food and water.
Karyotyping
To perform karyotyping on the iPSC clones, we first arrested the cell cycle of iPSCs by treating them with 10 μg/mL of colcemid (Thermo Fisher Scientific, MA, USA) for 20 min at 37 °C. We then incubated the cells with 75 mM hypotonic KCl, which led to nuclear swelling. Next, the cells were fixed using a 3:1 methanol/glacial acetic acid solution. Metaphase chromosomes were collected and subjected to Giemsa staining to analyze the G-bands. We counted at least 20 metaphase spreads for cytogenetic analysis.
Reverse transcription-polymerase chain reaction (RT-PCR) and quantitative real-time reverse transcription PCR (qRT-PCR)
In brief, total RNA was isolated and purified with the GENEzol TriRNA Pure Kit with DNase I treatment (Geneaid, Taiwan), and the cDNAs were synthesized with the SuperScript™ III Reverse Transcriptase (Thermo Fisher Scientific, MA, USA) according to the manufacturer’s instructions. RT-PCR was conducted using 2X SuperRed PCR Master Mix (BIOTOOLS Co., Ltd, Taiwan) according to the manufacturer’s instructions. A 20 μL solution containing 5 ng total cDNA was used for PCR in the following conditions: 95 °C 3 min; 35 cycles of 95 °C 30 s; 50–60 °C 30 s; and 72 °C 1–5 min. They were kept at 4 °C until electrophoresis using 1–2% agarose gel (depending on the size of the band). qRT-PCR was performed in 25 μL of reaction solution containing 5 ng RNA equivalent cDNA template, 400 nM of each primer, and 1X SYBR Green (Thermo Fisher Scientific, MA, USA). The PCR reactions were set up using a QuantStudio™ 5 Real-Time PCR System (Thermo Fisher Scientific, MA, USA). RNA levels were normalized to the GAPDH level and calculated as the delta-delta threshold cycle (ΔΔCT). Four replicate datasets were analyzed for each gene.
Alkaline phosphatase staining
Alkaline phosphatase staining was performed using the VECTOR Blue Alkaline Phosphatase Substrate kit (Vector Laboratories, CA, USA) according to the manufacturer’s instructions. Microscopic images were acquired by ZEISS Axio Observer 7 (Carl ZEISS, Jena, Germany).
IF staining
The samples were fixed in 4% paraformaldehyde for 20 min at room temperature, permeabilized with 0.1% Triton X-100 (5 min), and blocked in an FBS-BSA blocking buffer for 1 h at room temperature and incubated with the primary antibodies overnight at 4 °C. We also used Alexa 488/594-conjugated secondary antibodies (Biolegend, CA, USA). DAPI was used to stain the nuclei. The fluorescence microscopic images were acquired using ZEISS Axio Observer 7 (Carl ZEISS, Jena, Germany) and analyzed using ImageJ software (Fiji edition, National Institutes of Health, USA).
Generation of IDS mutation-corrected isogenic control (ISO) iPSCs
Base editing of the mutated IDS gene in MPS II-iPSCs derived from Patient 1 and Patient 4 was performed by transfecting cells (Pt1-4 and Pt 4-15 clones) with ABE8e-NG HiFi vectors (Gene Knockout/In Cell Line Modeling Core at the National Taiwan University College of Medicine, Taiwan), which contained Cas9 H840 nickase and sgRNA sequences specific to exon 8 or exon 9 of the IDS gene. The transfection was done using the transIT-LT1 reagent (Mirus Bio, WI, USA) according to the manufacturer’s instructions. Single-cell clones (corrected clones) were obtained by antibiotic selection and validated by Sanger sequencing and Edit R [79].
Neuronal differentiation of iPSCs
The neuronal differentiation of iPSCs was performed as previously described [80]. Briefly, iPSCs were dissociated using Accutase (FoliBio, Taiwan) and seeded on Matrigel-coated plates supplemented with StemFlex medium. The cells were then cultured in 3 N medium (DMEM/F12, neurobasal medium, non-essential amino acids, sodium pyruvate, Glutamax; penicillin-streptomycin, B27, N2, 2-mercaptoethanol, all from Thermo Fisher Scientific, MA, USA) containing 10 μM SB431542 (Selleckchem, TX, USA) and 1 μM Dorsomorphin (Cayman Chemical, MI, USA) for 12 days. The formed neuroepithelial clumps were digested by dispase, reseeded on Matrigel-coated plates, and incubated in 3 N medium containing 20 ng/ml bFGF (Thermo Fisher Scientific, MA, USA) for four days. Next, the cells were maintained in a 3 N medium without bFGF, which was replaced every three days until used for the experiments. After four weeks of differentiation, TUJ1 expression was detected by flow cytometry (FACS Calibur, BD Biosciences, NJ, USA). Neuronal differentiation for more than four weeks was also performed using a 3 N medium without bFGF.
IDS activity determination
IDS activity was determined as previously described [81]. Briefly, iPSCs and the derived neurons were used to measure the activity of IDS enzymes. Homogenates were made in water by sonication in a Bioruptor Plus sonication device (Diagenode, NJ, USA). The protein concentration was determined using a BCA protein assay kit (Merck-Millipore, Darmstadt, Germany). To measure IDS activity 10 μL (10 μg) of proteins were first incubated with 10 μL of 4-methylumbelliferyl-α-iduronate 2-sulfate (Moscerdam Substrates, Oegsteest, Netherlands) at 37 °C for 4 h. Second, the reaction was added with 20 μL McIlvains’ phosphate/citric acid buffer and 5 μL lysosomal enzymes purified from bovine testis (LEBT-M2, Moscerdam Substrates, Oegsteest, Netherlands) solution and incubated at 37 °C for 24 h. Stop buffer (0.5 M NaHCO3/0.5 M Na2CO3 buffer, pH 10.7 + 0.025% Triton X-100) was then added to terminate the reaction, and the fluorescence of 4-methylumbelliferyl-α-iduronate 2-sulfate was measured using a SpectraMax i3x Multi-Mode Microplate reader (Molecular Devices, CA, USA).
XTT assay
The 4-week and 15-week control and MPS II neurons were seeded in 96-well Matrigel-coated plates and incubated with XTT labeling mixture (Cayman Chemical, MI, USA). After 2 h, the supernatant was pipetted into 96-well plates, and the conversion of XTT to formazan was quantified by measuring the spectral absorbance at 450 nm using a SpectraMax i3x Multi-Mode Microplate reader (Molecular Devices, CA, USA).
Cellular glycosaminoglycan content
Sulfated proteoglycans and GAGs were quantified using the 1,9-dimethylmethylene blue (DMMB; Sigma-Aldrich, MA, USA) protocol [82]. The iPSCs and neurons were digested in a prepared papain solution (containing 1 mM EDTA, 2 mM DTT, and 0.3 mg/mL papain enzyme) (Sigma-Aldrich, MA, USA) at 60 °C for 1 h. Iodoacetic acid (Sigma-Aldrich, MA, USA) was then added to a final concentration of 10 mM, followed by the addition of 0.5 ml of 50 mM Tris/HCl (Sigma-Aldrich, MA, USA). Briefly, 20 μL of sample was mixed with 200 μL of DMMB reagent, and absorbance was read at 525 nm. A standard curve was established from chondroitin-6-sulfate (Sigma-Aldrich, MA, USA) to compare absorbance for the samples. The DNA content was measured using a fluorometric assay. Total GAG levels were normalized to total DNA content. Aliquots of the sample digestion were stained with 200 μL of Hoechst33258 (Cayman Chemical, MI, USA) working solution (2 μg/mL). The fluorescence intensities were then detected at 355 nm for excitation and 460 nm for emission. Both Hoechst-stained DNA and GAG contents were measured by a SpectraMax i3x Multi-Mode Microplate reader (Molecular Devices, CA, USA).
Electrophysiology and neurite length detection
AAV8-hSyn-EGFP, AAV9-CaMKII α-EYFP, and AAV9-mDlx-GFP (titer: ~3 × 1013 genome copies per milliliter; Addgene, MA, USA) were used to label iPSC-derived neurons (14 weeks of differentiation) used in the electrophysiology study. Whole-cell patch-clamp recordings were performed when the neurons were differentiated for 16–17 weeks. The coverslips containing neurons were transferred to an immersion-type recording chamber mounted on an upright microscope (Axio Examiner D1, Carl ZEISS, Jena, Germany) and continuously perfused with oxygenated artificial cerebrospinal fluid consisting of 124 mM NaCl, 3 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 1.25 mM NaH2PO4, 26 mM NaHCO3 and 20 mM glucose (pH 7.4, 295 mOsmol) at a rate of 2–3 mL/min and maintained at 30–32 °C. Neurons were viewed using Nomarski optics. Patch-clamp recordings of iPSC-derived neurons were performed using patch pipettes pulled from borosilicate glass tubing (1.5-mm outer diameter, 0.32-mm wall thickness; Sutter, Novato, CA, USA), with a resistance of approximately 6–8 MΩ when filled with the internal solution containing 100 mM K-gluconate, 20 mM KCl, 0.2 mM EGTA, 10 mM HEPES, 4 mM ATP-Mg, 0.3 mM GTP-Na, and 0.05% neurobiotin (pH 7.2, 295 mOsmol) (NEUROBIOTIN Tracer, SP-1120, Vector Laboratories, Inc., Burlingame, CA). Recordings were performed in current-clamp mode using a patch amplifier (Multiclamp 700 B; Molecular Devices, CA, USA).
The internal solution contained 100 mM K-gluconate, 20 mM KCl, 0.2 mM EGTA, 10 mM HEPES, 4 mM ATP-Mg, 0.3 mM GTP-Na, and 0.05% neurobiotin (pH 7.2, 295 mOsmol). The bridge was balanced, and the current was applied to the neurons to reach and maintain a nominal membrane potential of −70 mV. Neurons were accepted for further analysis only if the action potential (AP) exceeded 0 mV. The AP threshold was determined when dVm/dt reached 10 V/s. The properties of a single AP were monitored from the first AP elicited by the step depolarizing protocol: intracellular current was injected from −50 to 180 in 10 pA steps. The AP spike height was measured from the threshold to the peak of the spike. After the AP spike, after-hyperpolarization (AHP) was determined as the voltage difference between the threshold and the negative voltage point. The half-width of the AP was measured as the width at a half-maximal spike. The 10–90% rising slope and 90–10% decay slope of AP were determined using Clampfit 10.6 software (Molecular Devices, CA, USA). For accurate neuron reconstruction directly from an automatically inverted fluorescence microscope, images were acquired by ZEISS Axiovert 200 M (Carl ZEISS, Jena, Germany) and analyzed using Neurolucida software (MBF Bioscience, VT, USA).
RNA sequencing and gene pathway analysis
The extracted total RNAs from the 15-week HC neurons (three clones) and MPS II neurons (nine clones: three clones derived from Patient 1 and two clones derived from each of Patients 2, 3, and 4) were subjected to quality control, RNA sequencing, and analyses using the BIOTOOLS (BIOTOOLS Co., Ltd, Taiwan) platform. Briefly, Trimmomatic software was used to filter and remove low-quality reads from the raw data, resulting in clean reads, which were then mapped to the GRCh38 reference genome using HISAT2. The featureCounts software was used to count the number of raw reads per gene. The read count matrix was normalized by methods that included Relative Log Expression, Trimmed Mean of M-values, and Fragments Per Kilobase of transcript per Million mapped reads to correct for differences in sequencing depth and gene length. Differential gene expression analysis was performed using either DESeq2 or DEGseq/edgeR. Genes with a |Fold Change| > 2 and corrected p value < 0.005 were considered differentially expressed. The Kyoto Encyclopedia of Genes and Genomes (KEGG), ingenuity pathway analysis (IPA), and gene set enrichment analysis (GESA) were used to analyze functional enrichment. The RNA-seq dataset in this study was deposited in the Gene Omnibus Database (GSE202344).
Western blotting
For total protein extraction, the cultured iPSC-derived neurons were lysed with RIPA buffer (50 mM Tris, 150 mM NaCl, 100 mM EDTA, 1% sodium deoxycholate, 0.1% Triton X-100, 0.1% SDS, pH 7.4) at −80 °C for 30 min and centrifuged at 13,000 × g for 5 min (4 °C). The cell lysates were then collected.
For the extraction of cytoplasmic and nuclear fractions, cultured iPSC-derived neurons were first lysed in cytoplasmic fraction buffer (10 mM Tris, 10 mM KCl, 1 mM MgCl2, 0.1% Triton X-100, pH 7.4) on ice for 30 min and centrifuged at 13,000 × g for at 4 °C for 5 min, and the cytoplasmic fractions were collected. The pellet was then washed with cytoplasmic fraction buffer and centrifuged at 100 × g at 4 °C for 10 min three times to remove the residual cytoplasmic fraction. The pellet was then lysed in radioimmunoprecipitation assay buffer, frozen at −80 °C for 30 min, and centrifuged at 13,000 × g at 4 °C for 5 min. The supernatants were then collected as nuclear fractions. Protein concentrations in the cell lysates were determined using a BCA Protein Assay Kit (Merck-Millipore, Darmstadt, Germany). Cell lysates containing 30 μg of protein per sample were electrophoresed on a sodium dodecyl sulfate-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane. The membrane was incubated with primary antibodies at 4 °C overnight, followed by washing with Tris-buffered saline with 0.1% Tween 20 three times and incubation with horseradish peroxidase-conjugated appropriate secondary antibodies for 1 h at room temperature. The proteins were detected using an ECL Western blotting reagent (RPN2235; Cytiva, MA, USA) with an image analyzer (FUSION Solo S, Vilber, Paris, France). The intensities of the bands were quantified using ImageJ (National Institutes of Health, USA).
Flow cytometry analysis and fluorescence-activated cell sorting (FACS) sorting
Cells were fixed with 4% paraformaldehyde for 20 min at room temperature. Depending on the antibody used, cells were permeabilized with either 0.1% Triton X-100 (for most antibodies) or 0.1% saponin (specifically for LC3B staining) for 5 min. After blocking in FBS-BSA buffer for 1 h at room temperature, cells were incubated with primary antibodies overnight at 4 °C. This was followed by an incubation with Alexa 488/594-conjugated secondary antibodies (Biolegend, CA, USA). Flow cytometry analyses were conducted on a FACSCalibur cytometer (BD Biosciences, NJ, USA), with data acquisition using CellQuest software. For cell sorting, a FACSAria IIIu instrument (BD Biosciences, NJ, USA) was utilized. In all cases, a minimum of 10,000 cells per condition were assessed, using linear gains for light scatter channels and logarithmic gains for fluorescence channels. Data were analyzed using FlowJo software (BD Biosciences, NJ, USA).
Luciferase reporter assay
The iPSC-derived 15-week neurons were infected with the pLAS5w-GFP-Puro-TCF virus. GFP-positive cells with equal mean fluorescent intensity were sorted and seeded on a 96-well plate by FACSAria III (BD Biosciences, NJ, USA), and luciferase activity was analyzed using a luciferase assay kit (Promega Luciferase Assay System, Promega, WI, USA).
Assays for cell viability
Membrane-permeable dyes calcein acetoxymethyl ester (calcein-AM; Biolegend, CA, USA) was prepared as stock solutions of 1 mM stored at −20 °C. The cells were divided into two tubes and with or without treated with calcein-AM (1 μM, 20 min) at 37 °C to assay the degrees of viability, respectively. The results were analyzed by flow cytometry, as described above.
ChIP analysis
The 17-week-old MPS II neurons were first crosslinked with 1% formaldehyde solution, followed by reverse crosslinking with 0.125 M glycine solution. The cells were then lysed on ice for 15 min in ChIP lysis buffer and sonicated for 30 sec on and 30 sec off in five cycles using a Bioruptor Plus sonication device (Diagenode, NJ, USA). The isolated 200–1000 base pairs of DNA fragments were immunoprecipitated with anti-non-phosphorylated β-catenin antibody (Cell Signaling, MA, USA) at 4 °C overnight, followed by incubation with 50 µl of protein G (Cytiva, MA, USA) at 4 °C for 4 h. The recovery of DNA fragments was achieved by reversing the crosslinking process using 5 mM NaCl and 40 µg RNaseA at 65 °C overnight, followed by 4 µl (10 mg/ml) of proteinase K (Qiagen Inc., CA, USA) at 60 °C for 1 h. The immunocomplex was purified by ethanol precipitation after being washed and eluted from beads. The human SCN9A promoter region was detected using PCR and quantified using qRT-PCR.
Intracellular calcium measurements
The intracellular calcium levels in neurons were measured either by calcium imaging or microplate reader method. The calcium imaging was performed utilizing a Nikon Ti microscope equipped with a Qi2 camera and a CFI Plan Apochromat Lambda D objective lens (Nikon, Japan). Alternatively, the fluorescence from neurons in flat-bottom black microplates (Thermo Fisher Scientific) was detected using a SpectraMax i3x Multi-Mode Microplate reader (Molecular Devices, CA, USA). To start the experiment, neurons (3.5 × 104 cells/well) were cultured in 96-well Clear (for calcium imaging) or Black (for microplate reader method) Bottom Plates (Thermo Fisher Scientific) and were loaded with 4 µM Fura-2, 0.08% Pluronic™ F-127, and 1 mM probenecid (all from Thermo Fisher Scientific) in extracellular buffer (ECB), which includes 125 mM NaCl, 5 mM KCl, 1.5 mM MgCl2, 20 mM HEPES, 10 mM Glucose, and 1.5 mM CaCl2, for 30 min at room temperature. The dye was then washed out in ECB, and cells were incubated for an additional 30 min to allow for complete de-esterification of the dye. Baseline intracellular calcium levels were measured at 340 nm and 380 nm wavelengths either by microscopy or a microplate reader over a two-min period. In microscopy, the mean calcium fluorescence levels were quantified from 50 neurons per time point with ImageJ software. A minimum of four wells were utilized for biological replicates. Subsequently, neurons were treated with 4 µM Thapsigargin and 3 mM EGTA (both from Thermo Fisher Scientific) to induce immediate calcium shifts. Post-treatment images were captured at both 340 nm and 380 nm wavelengths over a 9-min period, while the post-treatment fluorescence was alternatively measured at the same wavelengths using a microplate reader over a 15-min period. The fluorescence intensity ratios at 340 nm and 380 nm (F340/F380) were then obtained from both methods. The differences between post-treatment and baseline F340/F380 ratios, represented as “delta F340/F380”, were calculated. The integrals of the delta F340/F380 ratio plotted against time were computed to illustrate changes in intracellular calcium levels using GraphPad Prism 8 Software (GraphPad Software, CA, USA).
Statistical analysis
All data were presented as mean plus standard error of the mean (SEM), with the number of biological replicates (n) shown in figure legends. The Mann–Whitney U test, unpaired or paired Student’s t test, Wilcoxon signed-rank test, Welch’s t test, and chi-square test were performed using GraphPad Prism 8 Software (GraphPad Software). P values are reported as follows: ns, nonsignificant; *p < 0.05; **p < 0.01; ***p < 0.005.
Data availability
The RNA sequencing data in this study were deposited in the Gene Expression Omnibus (GEO): GSE202344. Please visit and enter the token “azmvwuaglpetlgd”.
References
Coutinho MF, Lacerda L, Alves S. Glycosaminoglycan storage disorders: a review. Biochem Res Int. 2012;2012:471325.
Hunter C. A Rare Disease in Two Brothers. Proc R Soc Med. 1917;10:104–16.
Demydchuk M, Hill CH, Zhou A, Bunkóczi G, Stein PE, Marchesan D, et al. Insights into Hunter syndrome from the structure of iduronate-2-sulfatase. Nat Commun. 2017;8:15786.
Muenzer J, Hendriksz CJ, Fan Z, Vijayaraghavan S, Perry V, Santra S, et al. A phase I/II study of intrathecal idursulfase-IT in children with severe mucopolysaccharidosis II. Genet Med. 2016;18:73–81.
D’Avanzo F, Rigon L, Zanetti A, Tomanin R. Mucopolysaccharidosis Type II: One Hundred Years of Research, Diagnosis, and Treatment. Int J Mol Sci. 2020;21:1258.
Caccamo D, Katsetos CD, Herman MM, Frankfurter A, Collins VP, Rubinstein LJ. Immunohistochemistry of a spontaneous murine ovarian teratoma with neuroepithelial differentiation. Neuron-associated beta-tubulin as a marker for primitive neuroepithelium. Lab Invest. 1989;60:390–8.
Holper S, Watson R, Yassi N. Tau as a biomarker of neurodegeneration. Int J Mol Sci. 2022;23:7307.
Carden MJ, Trojanowski JQ, Schlaepfer WW, Lee V. Two-stage expression of neurofilament polypeptides during rat neurogenesis with early establishment of adult phosphorylation patterns. J Neurosci. 1987;7:3489–504.
Schimmelpfeng J, Weibezahn K-F, Dertinger H. Quantification of NGF-dependent neuronal differentiation of PC-12 cells by means of neurofilament-L mRNA expression and neuronal outgrowth. J Neurosci Methods. 2004;139:299–306.
Lorenzo DN. Cargo hold and delivery: Ankyrins, spectrins, and their functional patterning of neurons. Cytoskeleton. 2020;77:129–48.
Catterall WA, Goldin AL, Waxman SG. International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev. 2005;57:397–409.
Yu M, Guo L, Li N, Henzel KS, Gu H, Ran X, et al. Overexpression of Kcnmb2 in dorsal CA1 of offspring mice rescues hippocampal dysfunction caused by a methyl donor-rich paternal diet. Front Cell Neurosci. 2018;12:360.
Johnston J, Forsythe ID, Kopp‐Scheinpflug C. SYMPOSIUM REVIEW: Going native: voltage‐gated potassium channels controlling neuronal excitability. J Physiol. 2010;588:3187–200.
Li N, Gao W, Zhang Y-F, Ho M. Glypicans as cancer therapeutic targets. Trends Cancer. 2018;4:741–54.
Xiong X, Chorzalska A, Dubielecka PM, White JR, Vedvyas Y, Hedvat CV, et al. Disruption of Abi1/Hssh3bp1 expression induces prostatic intraepithelial neoplasia in the conditional Abi1/Hssh3bp1 KO mice. Oncogenesis. 2012;1:e26.
Sadras T, Perugini M, Kok CH, Iarossi DG, Heatley SL, Brumatti G, et al. Interleukin-3-mediated regulation of β-catenin in myeloid transformation and acute myeloid leukemia. J Leukoc Biol. 2014;96:83–91.
Kleszcz R, Szymańska A, Krajka-Kuźniak V, Baer-Dubowska W, Paluszczak J. Inhibition of CBP/β-catenin and porcupine attenuates Wnt signaling and induces apoptosis in head and neck carcinoma cells. Cell Oncol. 2019;42:505–20.
Sobolev VV, Khashukoeva AZ, Evina OE, Geppe NA, Chebysheva SN, Korsunskaya IM, et al. Role of the transcription factor FOSL1 in organ development and tumorigenesis. Int J Mol Sci. 2022;23:1521.
Pellicano F, Thomson RE, Inman GJ, Iwata T. Regulation of cell proliferation and apoptosis in neuroblastoma cells by ccp1, a FGF2 downstream gene. BMC Cancer. 2010;10:1–12.
Partanen J. FGF signalling pathways in development of the midbrain and anterior hindbrain. J Neurochem. 2007;101:1185–93.
Musa A, Tripathi S, Dehmer M, Emmert-Streib F. L1000 Viewer: A Search Engine and Web Interface for the LINCS Data Repository. Front Genet. 2019;10:557.
Liang W, Huang X, Chen W. The effects of baicalin and baicalein on cerebral ischemia: a review. Aging Dis. 2017;8:850.
Zhou A, Lin K, Zhang S, Chen Y, Zhang N, Xue J, et al. Nuclear GSK3β promotes tumorigenesis by phosphorylating KDM1A and inducing its deubiquitylation by USP22. Nat cell Biol. 2016;18:954–66.
Entchev E, Jantzen I, Masson P, Bocart S, Bournique B, Luccarini JM, et al. Odiparcil, a potential glycosaminoglycans clearance therapy in mucopolysaccharidosis VI-Evidence from in vitro and in vivo models. PLoS One. 2020;15:e0233032.
Warford JR, Lamport A-C, Clements DR, Malone A, Kennedy BE, Kim Y, et al. Surfen, a proteoglycan binding agent, reduces inflammation but inhibits remyelination in murine models of Multiple Sclerosis. Acta Neuropathologica Commun. 2018;6:1–21.
Vallarola A, Tortarolo M, De Gioia R, Iamele L, de Jonge H, de Nola G, et al. A novel HGF/SF Receptor (MET) agonist transiently delays the disease progression in an amyotrophic lateral sclerosis mouse model by promoting neuronal survival and dampening the immune Dysregulation. Int J Mol Sci. 2020;21:8542.
Huang W, Zhu PJ, Zhang S, Zhou H, Stoica L, Galiano M, et al. mTORC2 controls actin polymerization required for consolidation of long-term memory. Nat Neurosci. 2013;16:441–8.
Sonawane SK, Uversky VN, Chinnathambi S. Baicalein inhibits heparin-induced Tau aggregation by initializing non-toxic Tau oligomer formation. Cell Commun Signal. 2021;19:16.
Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–83.
Horgan C, Jones SA, Bigger BW, Wynn R. Current and Future Treatment of Mucopolysaccharidosis (MPS) Type II: Is Brain-Targeted Stem Cell Gene Therapy the Solution for This Devastating Disorder? Int J Mol Sci. 2022;23:4854.
Taylor M, Khan S, Stapleton M, Wang J, Chen J, Wynn R, et al. Hematopoietic Stem Cell Transplantation for Mucopolysaccharidoses: Past, Present, and Future. Biol Blood Marrow Transplant. 2019;25:e226–e246.
Kobolak J, Molnar K, Varga E, Bock I, Jezso B, Teglasi A, et al. Modelling the neuropathology of lysosomal storage disorders through disease-specific human induced pluripotent stem cells. Exp Cell Res. 2019;380:216–33.
Rybova J, Ledvinova J, Sikora J, Kuchar L, Dobrovolny R. Neural cells generated from human induced pluripotent stem cells as a model of CNS involvement in mucopolysaccharidosis type II. J Inherit Metab Dis. 2018;41:221–9.
Hong J, Cheng YS, Yang S, Swaroop M, Xu M, Beers J, et al. iPS-derived neural stem cells for disease modeling and evaluation of therapeutics for mucopolysaccharidosis type II. Exp Cell Res. 2022;412:113007.
Reboun M, Rybova J, Dobrovolny R, Vcelak J, Veselkova T, Storkanova G, et al. X-Chromosome Inactivation Analysis in Different Cell Types and Induced Pluripotent Stem Cells Elucidates the Disease Mechanism in a Rare Case of Mucopolysaccharidosis Type II in a Female. Folia Biol. 2016;62:82–9.
Varga E, Nemes C, Bock I, Varga N, Feher A, Dinnyes A, et al. Generation of Mucopolysaccharidosis type II (MPS II) human induced pluripotent stem cell (iPSC) line from a 1-year-old male with pathogenic IDS mutation. Stem Cell Res. 2016;17:482–4.
Yong Y, Hunter-Chang S, Stepanova E, Deppmann C. Axonal spheroids in neurodegeneration. Mol Cell Neurosci. 2021;117:103679.
Stokin GB, Lillo C, Falzone TL, Brusch RG, Rockenstein E, Mount SL, et al. Axonopathy and transport deficits early in the pathogenesis of Alzheimer’s disease. Science. 2005;307:1282–8.
Alavi Naini SM, Soussi-Yanicostas N. Heparan Sulfate as a Therapeutic Target in Tauopathies: Insights From Zebrafish. Front Cell Dev Biol. 2018;6:163.
Hampe CS, Yund BD, Orchard PJ, Lund TC, Wesley J, McIvor RS. Differences in MPS I and MPS II Disease Manifestations. Int J Mol Sci. 2021;22:7888.
Holmes BB, DeVos SL, Kfoury N, Li M, Jacks R, Yanamandra K, et al. Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc Natl Acad Sci USA. 2013;110:E3138–47.
Shapiro EG, Nestrasil I, Delaney KA, Rudser K, Kovac V, Nair N, et al. A Prospective Natural History Study of Mucopolysaccharidosis Type IIIA. J Pediatr. 2016;170:278–87.e1-4.
Ohmi K, Kudo LC, Ryazantsev S, Zhao HZ, Karsten SL, Neufeld EF. Sanfilippo syndrome type B, a lysosomal storage disease, is also a tauopathy. Proc Natl Acad Sci USA. 2009;106:8332–7.
Hernandez F, Lucas JJ, Avila J. GSK3 and tau: two convergence points in Alzheimer’s disease. J Alzheimers Dis. 2013;33:S141–4.
Chauhan N, Paliwal S, Jain S, Verma K, Paliwal S, Sharma S. GSK-3β and its Inhibitors in Alzheimer’s Disease: A Recent Update. Mini Rev Medicinal Chem. 2022;22:2881–95.
Bikkavilli RK, Feigin ME, Malbon CC. p38 mitogen-activated protein kinase regulates canonical Wnt–β-catenin signaling by inactivation of GSK3β. J Cell Sci. 2008;121:3598–607.
Thornton TM, Pedraza-Alva G, Deng B, Wood CD, Aronshtam A, Clements JL, et al. Phosphorylation by p38 MAPK as an alternative pathway for GSK3beta inactivation. Science. 2008;320:667–70.
Ehyai S, Dionyssiou MG, Gordon JW, Williams D, Siu KM, McDermott JC. A p38 Mitogen-Activated Protein Kinase-Regulated Myocyte Enhancer Factor 2–β-Catenin Interaction Enhances Canonical Wnt Signaling. Mol Cell Biol. 2016;36:330–46.
Toualbi K, Güller M, Mauriz J, Labalette C, Buendia M, Mauviel A, et al. Physical and functional cooperation between AP-1 and β-catenin for the regulation of TCF-dependent genes. Oncogene. 2007;26:3492–502.
He Y, She H, Zhang T, Xu H, Cheng L, Yepes M, et al. p38 MAPK inhibits autophagy and promotes microglial inflammatory responses by phosphorylating ULK1. J Cell Biol. 2018;217:315–28.
Ryu HY, Kim LE, Jeong H, Yeo BK, Lee JW, Nam H, et al. GSK3B induces autophagy by phosphorylating ULK1. Exp Mol Med. 2021;53:369–83.
Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12:781–90.
Qiang L, Wu C, Ming M, Viollet B, He YY. Autophagy controls p38 activation to promote cell survival under genotoxic stress. J Biol Chem. 2013;288:1603–11.
Bigger BW, Begley DJ, Virgintino D, Pshezhetsky AV. Anatomical changes and pathophysiology of the brain in mucopolysaccharidosis disorders. Mol Genet Metab. 2018;125:322–31.
Costa R, Urbani A, Salvalaio M, Bellesso S, Cieri D, Zancan I, et al. Perturbations in cell signaling elicit early cardiac defects in mucopolysaccharidosis type II. Hum Mol Genet. 2017;26:1643–55.
Salvalaio M, D’Avanzo F, Rigon L, Zanetti A, D’Angelo M, Valle G, et al. Brain RNA-Seq Profiling of the Mucopolysaccharidosis Type II Mouse Model. Int J Mol Sci. 2017;18:1072.
Petherick KJ, Williams AC, Lane JD, Ordonez-Moran P, Huelsken J, Collard TJ, et al. Autolysosomal beta-catenin degradation regulates Wnt-autophagy-p62 crosstalk. EMBO J. 2013;32:1903–16.
Billings PC, Pacifici M. Interactions of signaling proteins, growth factors and other proteins with heparan sulfate: mechanisms and mysteries. Connect Tissue Res. 2015;56:272–80.
De Pasquale V, Sarogni P, Pistorio V, Cerulo G, Paladino S, Pavone LM. Targeting Heparan Sulfate Proteoglycans as a Novel Therapeutic Strategy for Mucopolysaccharidoses. Mol Ther Methods Clin Dev. 2018;10:8–16.
Jenkins LM, Singh P, Varadaraj A, Lee NY, Shah S, Flores HV, et al. Altering the Proteoglycan State of Transforming Growth Factor beta Type III Receptor (TbetaRIII)/Betaglycan Modulates Canonical Wnt/beta-Catenin Signaling. J Biol Chem. 2016;291:25716–28.
Drenth JP, Waxman SG. Mutations in sodium-channel gene SCN9A cause a spectrum of human genetic pain disorders. J Clin Investig. 2007;117:3603–9.
Matta C, Lewis R, Fellows C, Diszhazi G, Almassy J, Miosge N, et al. Transcriptome‐based screening of ion channels and transporters in a migratory chondroprogenitor cell line isolated from late‐stage osteoarthritic cartilage. J Cell Physiol. 2021;236:7421–39.
Segal M. Calcium stores regulate excitability in cultured rat hippocampal neurons. J Neurophysiol. 2018;120:2694–705.
Johnson ME, Gores GJ, Uhl CB, Sill JC. Cytosolic free calcium and cell death during metabolic inhibition in a neuronal cell line. J Neurosci. 1994;14:4040–9.
Korkotian E, Schwarz A, Pelled D, Schwarzmann G, Segal M, Futerman AH. Elevation of intracellular glucosylceramide levels results in an increase in endoplasmic reticulum density and in functional calcium stores in cultured neurons. J Biol Chem. 1999;274:21673–8.
Lloyd-Evans E, Morgan AJ, He X, Smith DA, Elliot-Smith E, Sillence DJ, et al. Niemann-Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nat Med. 2008;14:1247–55.
Gomez L, Thiebaut PA, Paillard M, Ducreux S, Abrial M, Crola Da Silva C, et al. The SR/ER-mitochondria calcium crosstalk is regulated by GSK3beta during reperfusion injury. Cell Death Differ. 2015;22:1890.
Michael A, Haq S, Chen X, Hsich E, Cui L, Walters B, et al. Glycogen synthase kinase-3beta regulates growth, calcium homeostasis, and diastolic function in the heart. J Biol Chem. 2004;279:21383–93.
Sweeney HL, Bowman BF, Stull JT. Myosin light chain phosphorylation in vertebrate striated muscle: regulation and function. Am J Physiol. 1993;264:C1085–95.
Berggard T, Arrigoni G, Olsson O, Fex M, Linse S, James P. 140 mouse brain proteins identified by Ca2+-calmodulin affinity chromatography and tandem mass spectrometry. J Proteome Res. 2006;5:669–87.
Joensuu M, Wallis TP, Saber SH, Meunier FA. Phospholipases in neuronal function: A role in learning and memory? J Neurochem. 2020;153:300–33.
Dominguez JM, Fuertes A, Orozco L, del Monte-Millan M, Delgado E, Medina M. Evidence for irreversible inhibition of glycogen synthase kinase-3beta by tideglusib. J Biol Chem. 2012;287:893–904.
Lovestone S, Boada M, Dubois B, Hüll M, Rinne JO, Huppertz H-J, et al. A phase II trial of tideglusib in Alzheimer’s disease. J Alzheimers Dis. 2015;45:75–88.
Horrigan J, Gomes TB, Snape M, Nikolenko N, McMorn A, Evans S, et al. A phase 2 study of AMO-02 (Tideglusib) in congenital and childhood-onset myotonic dystrophy type 1 (DM1). Pediatr Neurol. 2020;112:84–93.
Tolosa E, Litvan I, Höglinger GU, Burn D, Lees A, Andrés MV, et al. A phase 2 trial of the GSK‐3 inhibitor tideglusib in progressive supranuclear palsy. Mov Disord. 2014;29:470–8.
Martinez-Gonzalez L, Gonzalo-Consuegra C, Gomez-Almeria M, Porras G, de Lago E, Martin-Requero A, et al. Tideglusib, a Non-ATP Competitive Inhibitor of GSK-3beta as a Drug Candidate for the Treatment of Amyotrophic Lateral Sclerosis. Int J Mol Sci. 2021;22:8975.
Cheng Z, Han T, Yao J, Wang K, Dong X, Yu F, et al. Targeting glycogen synthase kinase-3beta for Alzheimer’s disease: Recent advances and future Prospects. Eur J Med Chem. 2024;265:116065.
Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76.
Kluesner MG, Nedveck DA, Lahr WS, Garbe JR, Abrahante JE, Webber BR, et al. EditR: A Method to Quantify Base Editing from Sanger Sequencing. CRISPR J. 2018;1:239–50.
Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc. 2012;7:1836–46.
Voznyi YV, Keulemans JL, van Diggelen OP. A fluorimetric enzyme assay for the diagnosis of MPS II (Hunter disease). J Inherit Metab Dis. 2001;24:675–80.
Zheng CH, Levenston ME. Fact versus artifact: avoiding erroneous estimates of sulfated glycosaminoglycan content using the dimethylmethylene blue colorimetric assay for tissue-engineered constructs. Eur Cell Mater. 2015;29:224–36.
Acknowledgements
We appreciate the service of the Flow Cytometric Analyzing and Sorting Core, the imaging core, and the Gene Knockout/in Cell Line Modeling Core of the First Core Labs at the National Taiwan University College of Medicine. We also appreciate the technical support provided by the Microscopy Core Facility of the Department of Medical Research at National Taiwan University Hospital. We thank members of the core facility of the Graduate Institute of Brain and Mind Sciences for providing a fluorescent microscope. We thank Dr. Chih-Kuang Chuang at the Division of Genetics and Metabolism and the Department of Medical Research at MacKay Memorial Hospital for helping with GAG measurement. We thank members of the Department of Pediatrics at Mackay Memorial Hospital for collecting blood from MPS II patients. We thank the members of Dr. Li-Jen Lee’s lab for assisting us in using Neurolucida software. We thank Miss Yu-Chiao Lin for assisting us in calcium imaging. Finally, we thank Drs. Sung-Tsang Hsieh, Ming-Shyue Lee, and Yi-Cheng Chang for their valuable scientific suggestions. We thank the Human Disease iPSC Service Consortium, funded by the National Science and Technology Council (NSTC 112-2740-B-001-004), for providing cells.
Funding
This work was supported by the Ministry of Science and Technology (MOST), Taiwan (103-2321-B-002-95, 104-2314-B-002-10-MY3, 107-2320-B-002-58 -MY3, 108-2319-B-001-4, 109-2740-B-001-2, 110-2740-B-001-3, 110-2320-B002-060, and 111-2320-B-002-50-MY3).
Author information
Authors and Affiliations
Contributions
HPH and TYC conceived the project and designed the experiments. SPL and HYL supervised the study and provided suggestions. LJL, HSH, and FCT aided in methodology. TYC and DFH performed the experiments. HPH and TYC analyzed the data and wrote the manuscript. All authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
Blood was collected from patients after obtaining their parents’ written informed consent. The protocol was approved by the Research Ethics Committee of both the National Taiwan University Hospital and Mackay Memorial Hospital. The reference number of this protocol is: 201502044RIND. The study was performed in accordance with the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Alexei Verkhratsky
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, TY., Lin, SP., Huang, DF. et al. Mature neurons from iPSCs unveil neurodegeneration-related pathways in mucopolysaccharidosis type II: GSK-3β inhibition for therapeutic potential. Cell Death Dis 15, 302 (2024). https://doi.org/10.1038/s41419-024-06692-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-024-06692-9