Abstract
Tumor-associated macrophages of the M2 phenotype promote cancer initiation and progression. Importantly, M2 macrophage-derived exosomes play key roles in the malignancy of cancer cells. Here, we report that circTMCO3 is upregulated in ovarian cancer patients, and its high expression indicates poor survival. M2-derived exosomes promote proliferation, migration, and invasion in ovarian cancer, but these effects are abolished by knockdown of circTMCO3. Furthermore, circTMCO3 functions as a competing endogenous RNA for miR-515-5p to reduce its abundance, thus upregulating ITGA8 in ovarian cancer. miR-515-5p inhibits ovarian cancer malignancy via directly downregulating ITGA8. The decreased oncogenic activity of circTMCO3-silencing exosomes is reversed by miR-515-5p knockdown or ITGA8 overexpression. Exosomal circTMCO3 promotes ovarian cancer progression in nude mice. Thus, M2 macrophage-derived exosomes promote malignancy by delivering circTMCO3 and targeting the miR-515-5p/ITGA8 axis in ovarian cancer. Our findings not only provide mechanistic insights into ovarian cancer progression, but also suggest potential therapeutic targets.
Similar content being viewed by others
Introduction
Ovarian cancer is one of the most common and fatal gynecological malignancies1. Due to the lack of effective early diagnosis strategies, ovarian cancer is typically diagnosed at an advanced stage with low 5-year survival rate of 29%2. Platinum-based chemotherapy, such as cisplatin, is one of standard therapies for ovarian cancer3,4. However, cisplatin resistance is one of major causes of therapeutic failure. Therefore, elucidating the mechanisms underlying the malignancy of ovarian cancer is crucial for developing therapeutic strategies.
Macrophages are crucial immune cells that can phagocytize bacteria and infected cells, present antigens and trigger inflammation5, which are typically divided into classically activated M1 macrophages and alternatively activated M2 macrophages. Importantly, tumor-associated macrophages (TAMs) account for about 50% of tumor mass with M2-polarized phenotypes6,7. TAMs are abundant in patients with ovarian cancer8, and TAMs promote ovarian cancer progression via regulating immune escape, migration, invasion, and metastasis9. However, TAM-mediated regulation of malignancy of ovarian cancer cells remains largely unknown. One of important mechanisms for macrophages is to release exosomes10. Exosomes released by M2 macrophages enhance migration and invasion in colon cancer11. Exploring macrophage-derived exosome-mediated regulation of ovarian cancer is crucial for a better understanding of the pathogenesis of ovarian cancer.
Circular RNAs (circRNAs) are non-coding RNAs which are formed by back-splicing12 and enriched in exosomes13. Strikingly, emerging evidence has revealed key roles of exosomal circRNAs in various human cancers. Chen and colleagues found that exosome-derived circ_0051443 repressed the malignancy of hepatocellular carcinoma (HCC) cells and HCC progression by promoting BAK1 expression14. In ovarian cancer, exosomal circWHSC1 promoted MUC1 expression and peritoneal diffusion and adhesion, contributing to cancer metastasis15. Guan et al. found that circPUM1 facilitated ovarian cancer progression, and exosomal circPUM1 promoted metastasis16. Thus, understanding the mechanisms by which exosomal circRNAs regulate ovarian cancer progression is truly important. A previous study reported increased abundance of circ_0031017 derived from TMCO3 gene (also known as circTMCO3) in TAM-derived exosomes17, suggesting the potential roles of exosomal circ_0031017 derived from M2 macrophages in cancers. Additionally, circTMCO3 was upregulated, and it sponged miR-577 to enhance proliferation, migration, and invasion in gastric cancer18. Wang et al. reported that circTMCO3 was highly expressed in HCC, and circTMCO3 might represent potentially valuable diagnostic biomarkers for HCC19. However, to our knowledge, the roles of circTMCO3 in cancers remain largely unknown, especially in ovarian cancer. Therefore, we focused the function of M2-derived exosomal circTMCO3 in ovarian cancer.
One of action mechanisms of circRNAs is that circRNAs function as competitive endogenous RNAs (ceRNAs) to absorb microRNAs (miRNAs) and release inhibitory effects on downstream targets20. CircRNAs modulate tumor-related gene expression via sponging miRNAs. miR-515-5p works as a tumor suppressor to inhibit proliferation, migration, and metastasis21. Integrins are key regulators of cancer growth and metastasis22. Integrin subunit alpha 8 (ITGA8) promotes epithelial-mesenchymal transition and cell invasion in multiple myeloma23. However, their roles and interaction in ovarian cancer are unknown. We found that miR-515-5p had a targeted binding relationship with circTMCO3 and ITGA8, respectively. Therefore, we hypothesized that M2-derived exosomal circTMCO3 might regulate malignant behaviors in ovarian cancer via sponging miR-515-5p and upregulating ITGA8.
To summarize, our investigation aims to study the roles of M2 macrophage-derived exosomes carrying circTMCO3 in regulating the malignancy of ovarian cancer cells. We found that the increased circTMCO3 expression was associated the poor survival of patients with ovarian cancer. Further investigation demonstrated that M2 macrophage-derived exosomal circTMCO3 promoted the malignancy by targeting the miR-515-5p/ITGA8 axis in ovarian cancer. Our findings are beneficial for deepening understanding of the progression of ovarian cancer and providing potential exosome-based therapeutic strategies.
Results
Abundant M2 macrophages facilitate ovarian cancer cell proliferation, migration, and invasion
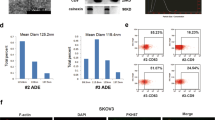
To investigate macrophage-mediated regulation of tumor progression, ovarian cancer tissues from patients were collected, and the M2 macrophage marker CD206 was detected via immunohistochemistry (IHC) staining. Compared to normal tissues, ovarian cancer tissues showed increased CD206-positive cells, suggesting increased abundance of M2 macrophages in ovarian cancer tissues (Fig. 1a). NTMs and TAMs were isolated, and the ratio of M2 macrophages (CD163 and CD206 double positive) was significantly increased in TAMs (Fig. 1b). Macrophages were isolated from tumor (TAM) and normal (NTM) tissues. Compared to NTMs, TAMs exhibited decreased the expression of M1 macrophage-related factors TNF-α and iNOS but increased the expression of M2 macrophage-associated factors Arg-1 and IL-10 (Fig. 1c). These data demonstrated that M2 macrophages were highly abundant in ovarian cancer. To explore whether M2 macrophages regulate the malignant behaviors of ovarian cancer cells, THP-1 cells were polarized to M2 macrophages, and A2780 and SKOV3 cells were cocultured with M0 or M2 macrophages. GW4869 was added to inhibit exosome generation. GW4869 significantly reduced total exosomal protein from M2 macrophages (Supplementary Fig. 1). Coculture with M2 macrophages greatly enhanced tumor cell proliferation, but it was abrogated by GW4869 (Fig. 1d, e). Furthermore, cell migration and invasion were enhanced by M2 macrophages, but these effects were largely suppressed by GW4869 treatment (Fig. 1f, g). These results suggest that abundant M2 macrophages promote malignancy in ovarian cancer cells via secreting exosomes.
a IHC staining of CD206 in ovarian cancer and normal tissues. Scale bar: 100 μm. b Flow cytometry analysis of CD163 and CD206-positive macrophages in NTMs and TAMs. c RT-qPCR analysis of TNFα, iNOS, IL-10 and Arg-1 in NTMs and TAMs. A2780 and SKOV3 cells were cultured with M0 or M2 macrophages. GW4869 was used to block exosome generation. d, e Cell proliferation was examined via CCK-8 and EdU incorporation assays (n = 3). Scale bar: 100 μm. f, g Migration and invasion were determined via wound healing and transwell assays (n = 3). Scale bar: 250 μm or 50 μm. *P < 0.05, **P < 0.01 and ***P < 0.001. Data were presented as mean ± standard deviation.
CircTMCO3 is upregulated in ovarian cancer and M2 macrophage-derived exosomes can be internalized by ovarian cancer cells
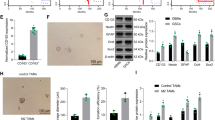
We isolated exosomes from TAMs and NTMs and examined the expression of hsa_circ_0000772, hsa_circ_0084335, hsa_circ_0031017 (circTMCO3), hsa_circ_0062531, hsa_circ_0017992, hsa_circ_0091577, and hsa_circ_0065492 in TAMs, NTMs and exosomes. hsa_circ_0000772, hsa_circ_0084335, hsa_circ_0031017 (circTMCO3) and hsa_circ_0091577 were upregulated in both TAMs and exosomes from TAMs, and hsa_circ_0031017 showed highest expression in TAMs (Supplementary Fig. 2a). Moreover, we examined the expression of circTMCO3 in THP-1 cells, M0 and M2 macrophages and found that circTMCO3 was highly expressed in M2 macrophages derived from THP-1 cells (Supplementary Fig. 2b). In addition, Northern blot assays showed that, compared to exosomes derived from NTMs, THP-1 cells and M0 macrophages, exosomes from TAMs and M2 macrophages derived from THP-1 cells showed increased circTMCO3 expression, respectively (Supplementary Fig. 2c). As circTMCO3 was abundant in cancer and TAM-derived exosomes, we analyzed circTMCO3 expression in ovarian cancer tissues from patients. CircTMCO3 expression was increased, and patients with high circTMCO3 expression exhibited poor survival (Fig. 2a, b). Strikingly, coculture with M2 macrophages upregulated circTMCO3 in A2780 and SKOV3 cells, and GW4869 treatment blocked circTMCO3 upregulation (Fig. 2c), indicating that M2 macrophages might enhance circTMCO3 expression through exosomes. As illustrated in Fig. 2d, circTMCO3 is generated from the exons 2–8 of TMCO3 on chromosome 13 with a length of 1423 nucleotides (nt) through back-splicing, and the junction site was verified by Sanger sequencing. Circular RNAs are quite stable in response to actinomycin D and RNase R treatment24,25. We found that, compared to linear TMCO3 mRNA, circTMCO3 exhibited decreased RNA decay after actinomycin treatment and high resistance to RNase R digestion (Fig. 2e, f). In addition, nuclear and cytoplastic fractions were prepared, and circTMCO3 could be detected in both nuclear and cytoplastic fractions, but it primarily localized in the cytoplasm (Fig. 2g). GW4869 suppressed circTMCO3 expression and M2-meditated oncogenic activity, suggesting that exosomes generated by M2 macrophages might be implicated in regulating circTMCO3 expression and ovarian cancer progression. Exosomes were then examined by transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA). Distinctive round bilayer vesicles with mainly 30–150 nm in diameter were observed (Fig. 2h, i). Moreover, M2 macrophage-derived exosomes showed high levels of extracellular vesicle proteins including tetraspanins (CD63, CD9, CD81) and TSG10126 compared to M2 macrophages (Fig. 2j). In coculture assays, we found that PKH26-labeled exosomes were internalized by ovarian cancer cells (Fig. 2k). The ratio of PKH26-positive cells was ~30% at 12 h, and it was increased to ~65% at 24 and 48 h (Fig. 2k). These results suggest that M2 macrophages raise circTMCO3 level in cancer cells via exosomes.
a CircTMCO3 expression was determined by RT-qPCR in patient tissues. b Patient survival. c RT-qPCR analysis of circTMCO3 in A2780 and SKOV3 cells in coculture assays (n = 3). d Genomic loci and the junction site of circTMCO3. Sanger sequencing was applied to confirm the junction site. e, f The abundance of circTMCO3 and linear TMCO3 mRNA in SKOV3 in response to actinomycin D and RNase R treatment was determined through RT-qPCR (n = 3). g The abundance of circTMCO3, U6 and GAPDH in nuclear and cytoplasmic fractions from SKOV3 cells was examined by RT-qPCR (n = 3). h The examination of M2 macrophage-derived exosomes using TEM. Scale bar: 200 nm. i Exosome size was evaluated by NTA. j Protein levels of CD63, CD9, CD81 and TSG101 in M2 macrophages and exosomes were analyzed by western blotting. k Exosomes (Red) were labeled with PKH26 and endocytosed by A2780 and SKOV3 cells (Nuclei, blue) after 12, 24 and 48 h. Scale bar: 25 μm. *P < 0.05, **P < 0.01 and ***P < 0.001. Data were presented as mean ± standard deviation.
M2 macrophage-derived exosomes promote the malignancy by delivering circTMCO3
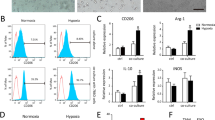
We proposed that M2 macrophage-derived exosomes might exert functions by delivering circTMCO3. Thus, circTMCO3 expression was knocked down by transfection of shcircTMCO3#1 or #2 in M2 macrophages, and M2-derived exosomes showed decreased circTMCO3 expression (Fig. 3a). A2780 and SKOV3 cells cocultured with Exo or Exo-shNC showed elevated expression of circTMCO3, but Exo-shcircTMCO3#1 and #2 had no significant effect on the expression of circTMCO3 in ovarian cancer cells (Fig. 3b), suggesting that M2 macrophage-derived exosomes delivered circTMCO3 into ovarian cancer cells. We found that Exo or Exo-shNC significantly raised 5-ethynyl-2′-deoxyuridine (EdU)-positive cell rate, migration, and invasion (Fig. 3c–e). However, Exo-shcircTMCO3#1 and #2 had no significant effect on the proliferation, migration, and invasion of ovarian cancer cells (Fig. 3c–e). Furthermore, compared to controls, A2780 and SKOV3 cells cocultured with Exo or Exo-shNC exhibited increased the expression of MMP9, Vimentin and Snail but decreased the expression of E-cadherin that were highly associated with cancer invasiveness and metastasis27,28,29,30, but these effects were abolished by Exo-shcircTMCO3#1 or #2 coculture (Fig. 3f). Collectively, these observations indicate that M2 macrophage-derived exosomal circTMCO3 enhances ovarian cancer cell proliferation, migration, and invasion. We further evaluated whether shcircTMCO3#1 and #2 reprogramed macrophages. Compared to Control, M2 and shNC, shcircTMCO3#1 and #2 significantly reduced CD163-positive M2 macrophages (Supplementary Fig. 3a). Moreover, shcircTMCO3#1 and #2 repressed the expression of M2-related markers including IL-10, Arg-1, Fizz-1 and TGF-β in M2 macrophages (Supplementary Fig. 3b–f), suggesting that inhibition of circTMCO3 suppressed M2 macrophage polarization.
a RT-qPCR analysis of circTMCO3 in M2 macrophages transfected with shNC, shcircTMCO3#1 or #2 (n = 3). A2780 and SKOV3 cells were cocultured with Exo, Exo-shNC, Exo-shcircTMCO3#1 or Exo-shcircTMCO3#2. b RT-qPCR analysis of circTMCO3 (n = 3). c EdU (red) incorporation analysis (n = 3; Scale bar, 100 µm; Nuclei, blue). d Wound healing assays for cell migration analysis (n = 3). Scale bar: 250 μm. e Transwell assays for cell invasion analysis (n = 3). Scale bar: 50 μm. f Protein levels of MMP9, E-cadherin, Vimentin and Snail were detected via western blotting. *P < 0.05, **P < 0.01 and ***P < 0.001. Data were presented as mean ± standard deviation.
Exosomal circTMCO3 functions as a ceRNA for miR-515-5p in ovarian cancer
As circRNAs regulate the expression of target genes via sponging miRNAs, a potential miR-515-5p binding site in circTMCO3 was predicted through Circinteractome (Fig. 4a). Ovarian cancer tissues showed low miR-515-5p expression, and miR-515-5p expression was negatively correlated with circTMCO3 expression (Fig. 4b, c). Besides, miR-515-5p was downregulated by Exo and Exo-shNC in A2780 and SKOV3 cells, but Exo-shcircTMCO3#1 and #2 lost the suppressive capacity (Fig. 4d). FISH assays showed colocalization of circTMCO3 and miR-515-5p in the cytoplasm (Fig. 4e). Furthermore, we found that miR-515-5p could be efficiently enriched by the circTMCO3 probe (Fig. 4f), and both circTMCO3 and miR-515-5p were enriched by an anti-Ago2 (Fig. 4g). The luciferase activity of circTMCO3-WT was inhibited by miR-515-5p overexpression but reinforced by miR-515-5p knockdown in ovarian cancer cells (Fig. 4h). However, neither overexpression nor knockdown of miR-515-5p affected the luciferase activity of circTMCO3-MUT (Fig. 4h). These data demonstrate that circTMCO3 acts as a miR-515-5p sponge to reduce its expression in ovarian cancer.
a The hsa-miR-515-5p binding site in circTMCO3. b RT-qPCR analysis of miR-515-5p in normal and tumor groups. c Pearson correlation analysis of circTMCO3 and miR-515-5p in tumor group. d RT-qPCR analysis of miR-515-5p in A2780 and SKOV3 cells in coculture assays (n = 3). e The colocalization of circTMCO3 and miR-515-5p were determined with FISH. Cells were hybridized with Alexa Fluor 488-labeled circTMCO3 (Green) and Cy3-labeled miR-515-5p (Red) probes, and the nuclei were stained with DAPI (Blue). Scale bar: 25 μm. f Direct binding of circTMCO3 and miR-515-5p was determined by RNA pull-down assays (n = 3). g The interaction of circTMCO3 and miR-515-5p was determined by RIP assays (n = 3). h Luciferase activity of wildtype (circTMCO3-WT) or mutant (circTMCO3-MUT) circTMCO3 reporters in A2780 and SKOV3 cells transfected with miR-515-5p mimics or inhibitor (n = 3). *P < 0.05, **P < 0.01 and ***P < 0.001. Data were presented as mean ± standard deviation.
miR-515-5p directly targets ITGA8 to inhibit its expression in ovarian cancer
To further explore downstream targets of miR-515-5p, we predicted a potential binding site for miR-515-5p in the 3′ untranslated region (UTR) of ITGA8 (Fig. 5a). Tumor tissues showed high expression of ITGA8 compared to normal tissues (Fig. 5b). ITGA8 expression was negatively correlated with miR-515-5p expression but positively correlated with circTMCO3 expression in ovarian cancer (Fig. 5c, d). Moreover, Exo and Exo-shNC significantly promoted ITGA8 expression, but it was abolished by knockdown of circTMCO3 (Fig. 5e). miR-515-5p was overexpressed or knocked down in A2780 and SKOV3 cells (Fig. 5f). ITGA8 was markedly reduced by miR-515-5p overexpression but elevated by miR-515-5p silencing (Fig. 5f). miR-515-5p overexpression impaired the luciferase activity of ITGA8-WT, but miR-515-5p knockdown raised its luciferase activity (Fig. 5g). The luciferase activity of ITGA8-MUT was not affected by transfection of miR-515-5p mimics or inhibitor (Fig. 5g). Therefore, miR-515-5p represses ITGA8 expression via binding to its 3′ UTR in ovarian cancer.
a The binding site for hsa-miR- 515-5p in the 3′ UTR of ITGA8. b RT-qPCR analysis of ITGA8 in patient tissues. c, d Pearson correlation analysis of ITGA8 and miR-515-5p or circTMCO3 in the tumor group. e RT-qPCR analysis of ITGA8 in A2780 and SKOV3 cells in coculture assays (n = 3). f The expression of miR-515-5p and ITGA8 in A2780 and SKOV3 cells transfected with miR-515-5p mimics, inhibitor, or negative controls (mimics and inhibitor NC) was determined via RT-qPCR (n = 3). g Luciferase activity of wildtype (ITGA8-WT) or mutant (ITGA8-MUT) ITGA8 reporters (n = 3). *P < 0.05, **P < 0.01 and ***P < 0.001. Data were presented as mean ± standard deviation.
miR-515-5p attenuates malignancy in ovarian cancer via downregulating ITGA8
To confirm whether miR-515-5p exerts anti-tumoral effects through ITGA8, ITGA8 was overexpressed in A2780 and SKOV3 cells through OE-ITGA8 transfection (Fig. 6a). Moreover, miR-515-5p mimics-mediated downregulation of ITGA8 was reversed by simultaneous overexpression of ITGA8 (Fig. 6b, c). miR-515-5p overexpression reduced EdU-positive cell rate and inhibited migration and invasion, but these suppressive effects were abrogated by overexpression of ITGA8 (Fig. 6d–f). The expression of MMP9, Vimentin and Snail were downregulated, and E-cadherin was upregulated in miR-515-5p-overexpressing cells, but simultaneous overexpression of ITGA8 reversed their expression (Fig. 6g). To conclude, miR-515-5p represses proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT) through suppression of ITGA8 in ovarian cancer.
a RT-qPCR analysis of ITGA8 in A2780 and SKOV3 cells transfected with OE-NC and OE-ITGA8 (n = 3). A2780 and SKOV3 cells were transfected with mimics NC, miR-515-5p mimics, miR-515-5p mimics in combination with OE-NC or OE-ITGA8. b, c RT-qPCR and western blotting analysis of ITGA8 (n = 3). d EdU incorporation analysis (n = 3). Scale bar: 100 μm. e Wound healing assays (n = 3). Scale bar: 250 μm. f Transwell assays (n = 3). Scale bar: 50 μm. g Protein levels of MMP9, E-cadherin, Vimentin and Snail were detected through western blotting. *P < 0.05, **P < 0.01 and ***P < 0.001. Data were presented as mean ± standard deviation.
Exosomal circTMCO3 promotes malignancy of ovarian cancer cells through the miR-515-5p/ITGA8 axis
We then examined whether exosomal circTMCO3-mediated regulation of malignancy in ovarian cancer was dependent on the miR-515-5p/ITGA8 axis. Exo-mediated acceleration of proliferation, migration, and invasion was suppressed by knockdown of circTMCO3 (Fig. 7a–c). Simultaneous knockdown of miR-515-5p or overexpression of ITGA8 reversed these effects and promoted proliferation, migration, and invasion (Fig. 7a–c). The expression of MMP9, Vimentin and Snail was enhanced, and E-cadherin was downregulated by Exo, but not by Exo-shcircTMCO3#1 and #2 (Fig. 7d). Knockdown of miR-515-5p or overexpression of ITGA8 reversed Exo-shcircTMCO3-mediated effects on these gene expressions (Fig. 7d). Therefore, exosomal circTMCO3 strengthens malignancy via targeting the miR-515-5p/ITGA8 axis.
A2780 and SKOV3 cells were transfected with miR-515-5p inhibitor or OE-ITGA8 and cocultured with Exo-shcircTMCO3#1 or Exo-shcircTMCO3#2. a EdU incorporation analysis (n = 3). Scale bar: 100 μm. b Wound healing assays (n = 3). Scale bar: 250 μm. c Transwell assays (n = 3). Scale bar: 50 μm. d Protein levels of MMP9, E-cadherin, Vimentin and Snail were detected via western blotting. *P < 0.05, **P < 0.01 and ***P < 0.001. Data were presented as mean ± standard deviation.
M2 macrophage-derived exosomal circTMCO3 accelerates ovarian cancer progression in vivo
We established an ovarian cancer xenograft mouse model via intraperitoneal injection, and mice were administrated with Exo, Exo-shNC, Exo-shcircTMCO3#1 or Exo-shcircTMCO3#2. Compared to control, Exo and Exo-shNC administration promoted tumor growth and raised tumor nodules and weight (Fig. 8a–c). However, Exo-shcircTMCO3#1 and #2 failed to exert these oncogenic effects (Fig. 8a–c). Moreover, we observed that Exo and Exo-shNC promoted the expression of circTMCO3 and ITGA8 and inhibited miR-515-5p expression, but it was abrogated by circTMCO3 knockdown (Fig. 8d–f). IHC staining showed that Exo and Exo-shNC administration enhanced the expression of Ki67, a widely used proliferation marker of cancer cells, and ITGA8 in tumors, and knockdown of circTMCO3 abrogated exosome-mediated upregulation of Ki67 and ITGA8 (Fig. 8g). Furthermore, ITGA8, MMP9, Vimentin and Snail were upregulated, and E-cadherin was downregulated by Exo and Exo-shNC administration, and these effects were suppressed by Exo-shcircTMCO3#1 and #2 (Fig. 8h). Collectively, these results demonstrate that M2 macrophage-derived exosomal circTMCO3 accelerates ovarian cancer progression in vivo.
Mice were intraperitoneally injected with 5 × 106 SKOV3 cells and administrated with control (PBS), Exo, Exo-shNC, Exo-shcircTMCO3#1 or Exo-shcircTMCO3#2. a Mouse abdomen was opened and imaged. b Tumor nodules were counted (n = 5). c Tumor weight (n = 5). d–f RT-qPCR analysis of circTMCO3, miR-515-5p and ITGA8 in tumors (n = 5). g IHC staining of Ki67 and ITGA8 in tumors. Scale bar: 100 μm. h Protein levels of ITGA8, MMP9, E-cadherin, Vimentin and Snail in tumors were examined by Western blotting. **P < 0.01 and ***P < 0.001. Data were presented as mean ± standard deviation.
Discussion
Ovarian cancer is a highly lethal cancer that accounts for 5% of all cancer-related deaths among women31,32. Ovarian cancer is generally diagnosed at a late stage, causing poor outcome of patients33. Therefore, exploring the regulatory mechanisms of ovarian cancer progression is very important. Here, we firstly demonstrated that M2 macrophage-derived exosomal circTMCO3 sponged miR-515-5p and relieved its inhibitor effect on ITGA8 expression, thereby promoting the malignancy and progression of ovarian cancer.
Exosomes play vital roles in cell-cell communication via transferring cell components and regulating behaviors of various cancer cells34,35. M2 macrophages attract much attention as the major component of tumor microenvironment. Exosomes released by M2 macrophages promote the metastasis of hepatocellular carcinoma cells and accelerate angiogenesis and growth of pancreatic cancer36,37. In consistence, we observed that M2 macrophages enhanced ovarian cancer cell proliferation, migration, and invasion via releasing exosomes. Exosomes have shown enormous potential in drug delivery with great biocompatibility and low immunogenicity38. Our data suggests that M2 macrophage-derived exosomes can be engineered for the management of ovarian cancer.
In recent years, exosomes delivering ncRNAs, such as lncRNAs, are becoming a pivotal regulator in regulating cancer progression39. Exosomal circRNAs have been regarded as potential therapeutic targets for various cancers40. Ma et al. found that exosomal circRNA051239 derived from tumors promoted proliferation, migration, and invasion of epithelial ovarian cancer cells via working as a miR-509-5p sponge and regulating PRSS3 expression41. In addition, circRNAs are key regulators of ovarian cancer progression and therapeutic targets for ovarian cancer42. However, our understanding of circRNAs and related regulatory mechanisms are still limited. CircTMCO3 exerts an oncogenic activity in gastric cancer, but its roles in other cancers including ovarian cancer remain unknown. Here, increased expression of circTMCO3 in M2 macrophages suggested us that exosomes might carry circTMCO3 to exert its function in ovarian cancer. Indeed, exosomes with knockdown of circTMCO3 lost their activity in regulating malignancy, demonstrating that exosomes exert their function via delivering circTMCO3 in ovarian cancer.
The ceRNA hypothesis suggests that lncRNAs work as miRNA sponges to upregulate downstream mRNA targets in cancers43. miR-577 has been identified as a target of circTMCO318. Wen et al. discovered that miR-515-5p repressed malignant behaviors in breast cancer through CBX444. miR-515-5p restrained migration and metastasis by downregulating MARK4 in lung and breast cancers45. However, the implication of miR-515-5p in ovarian cancer is unknown. Here, miR-515-5p was identified as a target of circTMCO3, and miR-515-5p restrained proliferation, migration, invasion, and EMT, discovering an anti-tumor activity in ovarian cancer.
The integrin family serves key roles in promoting cancer progression via facilitating proliferation, migration, invasion, and metastasis in various cancers46. As an integrin subunit, ITGA8 promotes EMT, migration, and invasion in multiple myeloma23, and its diagnostic and prognostic values in colon cancer have been validated47. However, its roles in ovarian cancer have never been reported previously. We showed ITGA8 as a downstream target of miR-515-5p in ovarian cancer. Our findings suggested that miR-515-5p reduced ITGA8 expression via binding to its 3′ UTR in ovarian cancer, and ITGA8 overexpression reversed miR-515-5p-mediated regulation of cancer malignancy and progression, indicating that miR-515-5p reduced ITGA8 expression to suppress proliferation, migration, invasion, and metastasis. However, ITGA8 is transmembrane receptor that mediates numerous cellular processes, and its implication in exosome internalization needs to be explored. Besides regulating ITGA8 expression, exosomal RNA-mediated regulation of ITGA8 function might be implicated in the pathogenesis of ovarian cancer.
Taken together, M2 macrophage-derived exosomes enhance malignancy in ovarian cancer by delivering circTMCO3 and targeting the miR-515-5p/ITGA8 axis. Our findings highlight exosomal circTMCO3-mediated regulation of malignancy of cancer cells and provide potential therapeutic strategies for ovarian cancer. Single cell sequencing will be performed in future studies to investigate the gene expression level difference between the cell types within the tumor and normal tissues.
Methods
Patients and specimens
Sixty patients were diagnosed with ovarian cancer at Hunan Cancer Hospital, and tumor and adjacent normal tissues were collected and stored for RNA extraction and immunohistochemistry staining. Inclusion criteria was following: 1 Greater than or equal to 18 years old; 2 First diagnosis with primary ovarian cancer; 3 No other severe diseases, such as cancers and diabetes; 4 No treatment; 5 No pregnancy and lactation. Macrophages were isolated from tumor (TAM) and normal (NTM) tissues and stained with CD11b and CD206 antibodies (101205 and 321105, Biolegend, San Diego, CA, USA). The cells were detected Beckman cytoflex flow cytometry (Beckman, USA), and the Cytexpert Software was applied for the flow cytometry data analysis. The gating strategy was provided in the Supplementary Fig. 4. Written informed consent was required from patients. The Ethics Committee of Hunan Cancer Hospital approved this study.
Cell culture and polarization
Human ovarian cancer cells, A2780 and SKOV3, were purchased from iCell Bioscience Inc (Shanghai, China) and Cell Bank, Chinese Academy of Sciences (Shanghai, China) and maintained in DMEM/10% FBS (Gibco, Waltham, MA, USA). Human monocytic cell THP-1 was bought from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were authenticated by STR profiling and tested for mycoplasma contamination. For macrophage polarization, THP-1 cells were induced with PMA (100 ng/mL, Sigma, St. Louis, MO, USA) for 48 h (M0) prior to additional 24-h incubation with LPS (100 ng/mL, Sigma) and IL-4 (20 ng/mL, PeproTech, Cranbury, NJ, USA) for M2 polarization. For 24-h coculture of macrophages and ovarian cancer cells, M0 or M2 macrophages (1 × 105) were plated on the upper chambers, and A2780 or SKOV3 cells (5 × 105) were seeded on the lower chambers. For GW4869 treatment, M2 macrophages were pretreated with GW4869 (10 μM, Sigma) for 8 h.
Cell transfection
For ITGA8 overexpression, the coding region of ITGA8 was cloned into pcDNA3.1 (OE-ITGA8, Invitrogen, Carlsbad, CA, USA). Mock transfection controls were prepared using the empty pcDNA3.1 vector (OE-NC). The shRNAs against circTMCO3 (shcircTMCO3#1 and #2), miR-515-5p mimics, inhibitor, and negative controls (shNC, mimics NC and inhibitor NC) were synthesized by RiboBio (Guangzhou, China). A2780 and SKOV3 cells were transfected using Lipo 3000 (Invitrogen) following the manual. For knockdown of circTMCO3, lentiviral particles were prepared and transduced into M2 macrophages. After 48 h, cells were harvested. The sequences of shcircTMCO3#1 and #2 were 5′-GTGCATCTTCTAGCTGAAAAT-3′ and 5′-GCATCTTCTAGCTGAAAATGT-3′.
Exosome extraction, characterization, and internalization
Exosomes were extracted from M2 macrophages using Total Exosome Isolation Reagent (Invitrogen). Culture media were centrifuged, and the supernatants were collected. The exosome isolation reagent was mixed with the supernatants thoroughly and incubated overnight at 4 °C. Samples were centrifuged, and the pellet was resuspended. Exosomes were quantified and co-cultured with A2780 or SKOV3 cells at 100 µg/mL for 48 h. TEM was applied to examine exosome morphology. exosome size was analyzed by NTA (Malvern, Westborough, MA, USA). Protein levels of CD9, CD63, CD81 and TSG101 were determined by western blotting. Exosomes were stained with PKH26 dye (Sigma) and incubated with A2780 and SKOV3 cells for 24 h for imaging.
CircTMCO3 characterization
SKOV3 cells were treated with actinomycin D (5 µg/mL, Sigma) for 0, 4, 8, 12, or 24 h, and RNA was extracted for analyzing the decay of circTMCO3 and TMCO3 mRNA. Moreover, RNA was isolated, treated with RNase R (5 U/µg, Abcam, Cambridge, UK) for 2 h and subjected RT-qPCR for examining the remaining of circTMCO3 and TMCO3 mRNA. CircTMCO3 was amplified using divergent primers, and the junction site was identified through Sanger sequencing (Sangon Biotech, Shanghai, China).
Nuclear and cytoplasmic fractionation
Nuclear/Cytosol Fractionation Kit was purchased from BioVision (Milpitas, CA, USA). Nuclear and cytoplasmic fractions were separated following the manual. The abundance of circTMCO3, U6 snRNA and GAPDH in nuclear and cytoplasmic fractions was examined by RT-qPCR.
Fluorescence in situ hybridization (FISH)
SKOV3 cells were seeded on coverslips and fixed in a mixture solution of methanol and acetic acid (3:1) for 10 min. Let coverslips dried naturally, and cells were immersed in hybridization solution containing Alexa Fluor 488-labeled circTMCO3 and Cy3-labeled miR-515-5p probes at 25 nM. Coverslips were denatured and incubated overnight. Coverslips were washed and mounted in antifade mountant with DAPI (Beyotime, Shanghai, China) for imaging.
Real-time quantitative reverse transcription-PCR (RT-qPCR)
RNA was isolated using Total RNA Isolation Kit (Thermo Fisher Scientific, Waltham, MA, USA) and reversely transcribed into cDNA. The mirPremier microRNA isolation kit (Sigma) was used to isolate miRNAs, which were reversely transcribed using the miScript kit (QIAGEN, Germantown, MD, USA). Quantitative PCR was applied to detect circTMCO3, TMCO3, miR-515-5p, ITGA8, TNF-α, iNOS, IL-10, Arg-1 using SYBR Green (Beyotime). GAPDH and U6 snRNA were used as normalization controls. The 2−∆∆Ct method was used to calculate their relative expression. Primers were listed in Supplementary Table 1.
Western blotting
Exosomal protein was isolated using Exosome RNA and Protein Isolation Kit (ThermoFisher Scientific). Tumor tissues were homogenized and A2780 and SKOV3 cells were suspended in Protein Extraction Buffer (Abcam) supplemented with protease inhibitors (Thermo Fisher Scientific), and the supernatants were collected after centrifugation. Protein concentration was determined with BCA Protein Assay Kit (Beyotime) according to the manufacturer’s suggestion. After quantification, protein was loaded at 30 µg each lane, electrophoresed, and transferred to polyvinylidene fluoride (PVDF) membranes (Bio-Rad, Hercules, CA, USA). The PVDF membranes were cropped according to the protein molecular weight recommended by the manufacturer’s instructions. Rabbit antibodies against CD9 (1:1000, ab223052), CD63 (1:2000, ab231975), CD81 (1:2000, ab155760), TSG101 (1:800, ab225877), MMP9 (1:500, ab283575), E-cadherin (1:1000, ab227639), Vimentin (1:500, ab137321), Sanil (1:500, ab216347), ITGA8 (1:1000, ab243027) and GAPDH (1:8000, ab9485) from Abcam (Cambridge, UK) were used to examine their protein levels. GAPDH was used a normalization control. An HRP-donkey anti-rabbit IgG antibody (A16023, ThermoFisher Scientific) was used to incubate membranes for 1 h. Protein bands were visualized using ECL substrate (Beyotime) and quantified with the ImageJ software. The original western blot images were presented Supplementary Fig. 5.
Northern blotting
Cells and exosomes were lysed, and total RNA was extracted and quantified by measuring the OD at 260 nm. Subsequently, RNA was separated by agarose gel electrophoresis and transferred to Nylon membrane. RNA was cross-linked to the membrane, prehybridized, and hybridized with the radiolabeled circTMCO3 probe. Then, the membrane was washed, and the signal was visualized using X-ray27.
Cell Counting Kit-8 (CCK-8) assay
After cell treatment, culture medium was removed, and 100 µL of medium was added. CCK-8 (10 µL, Sigma) was added and incubated for 4 h. The absorbance (450 nm) was recorded.
Wound healing assay
In brief, inserts were oriented in plates. A2780 and SKOV3 cells (5 × 105) were added and cultured at 37 °C. Then, inserts were removed, and cells were washed and cocultured with macrophages or exosomes as indicated. The wound healing process was monitored using the BX51 microscope from Olympus (Tokyo, Japan).
EdU incorporation assay
A2780 and SKOV3 cells were cocultured with exosomes as indicated in medium supplemented with EdU (10 µM, Thermo Fisher Scientific) for 24 h. Cells were fixed, permeabilized and processed for the Click-IT reaction following the manual. Cells were stained with DAPI (Beyotime) and captured using a confocal microscope (Nikon). The EdU positive rate (%) = the number of positive cells/total cells ×100%.
Transwell invasion assay
Cell invasion was assessed by transwell assays with transwell chambers (8-µm pore membranes, Corning, NY, USA) precoated with Matrigel (Corning). Cells (1 × 106 cells) were seeded on the upper chamber and cocultured with macrophages or exosomes as indicated. Cells which invaded into the lower chamber were fixed, stained with crystal violet (Sigma) and observed under the BX51 microscope (Olympus).
RNA immunoprecipitation (RIP)
RIP assays were performed using the RIP kit (Millipore, Burlington, MA, USA). Magnetic beads were coated with the rabbit anti-Ago2 (ab186733, Abcam) or normal IgG (2729, Cell Signaling Technology, Danvers, MA, USA). A2780 and SKOV3 cells were lysed, and lysates were harvested. Magnetic beads and lysates were mixed well and incubated overnight. RNA was recovered and subjected to RT-qPCR.
RNA pull-down
A2780 and SKOV3 cells were lysed and centrifugated for collecting cell lysates. Biotinylated circTMCO3 probe or control oligo probe was mixed with cell lysates and incubated for 16 h at 4 °C. Streptavidin magnetic beads (Thermo Fisher Scientific) were added, mixed well, and incubated for additional 2 h with gentle rotation. Subsequently, RNA was recovered, and the abundance of miR-515-5p was examined by RT-qPCR.
Dual-luciferase reporter assay
Wildtype and mutant binding sites for miR-515-5p in circTMCO3 (circTMCO3-WT and circTMCO3-MUT) and the 3′ untranslated region (3′ UTR) of ITGA8 (ITGA8-WT and ITGA8-MUT) were inserted into pmirGLO (Promega, Madison, WI, USA). A2780 and SKOV3 cells were cotransfected with luciferase reporters and miR-515-5p mimics or inhibitor. Cells were collected for determining the luciferase activity with the Dual-Glo Luciferase Assay System (Promega).
An intraperitoneal xenograft mouse model of ovarian cancer
Twenty female BALB/c nude mice (five-week-old, Hunan SJA Laboratory Animal, Changsha, China) were randomly divided into four groups (n = 5 per group): Control, Exo, Exo-shNC, Exo-shcircTMCO3#1 and Exo-shcircTMCO3#2. SKOV3 cells (1 × 106 cells in 200 µL of PBS) were injected intraperitoneally into nude mice. Subsequently, exosomes (30 µg in 200 µL of PBS) from M2, shNC or shcircTMCO3-transfected M2 macrophages were intraperitoneally injected into mice every three days. Control mice received 200 µL of PBS without exosomes. Mice were sacrificed through deep anesthesia with sodium pentobarbital after 21 days. The investigator was blinded to the group allocation during the experiment. Animal procedures were approved by the Animal Care and Use Committee of Hunan Cancer Hospital.
Immunohistochemistry (IHC) staining
Tumors from mice and ovarian cancer and normal adjacent tissues were process for paraffin embedding and cut into 5-µm slices. Antigen retrieval was performed in pH 6.0 antigen retrieval solution (Invitrogen), and slices were blocked consecutively in H2O2 and BSA solution. Slides were incubated with anti-CD206 (1:50, ab64693, Abcam), anti-Ki67 (1:50, ab833, Abcam) or ITGA8 (1:100, ab243027, Abcam) overnight. Subsequently, slides were washed and incubated with a donkey anti-rabbit or mouse HRP-conjugated secondary antibody, and DAB substrate (Beyotime) was added. Slides were stained with hematoxylin and imaged with the BX51 microscope (Nikon).
Statistics and reproducibility
Data from three independent experiments were presented as mean ± standard deviation. Kaplan–Meier curve was applied to analyze patient survival, and the survival time was statistically compared using the log rank test. The Pearson correlation was used to for correlation analysis of gene expression. A priori power analysis (G*Power software) was performed to assess sample size required to generate 80% power for detecting a significant (P < 0.05) effect of treatment. The normality of data was evaluated by the Shapiro–Wilk test. Considering the significance level of 5%, there were no significant deviations from the normality of all data (P > 0.05). The variance of groups was analyzed by the Student’s t test (two tailed) and one-way analysis of variance (ANOVA) with Turkey post hoc test. P < 0.05 was statistically significant. *P < 0.05, **P < 0.01 and ***P < 0.001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated or analyzed during this study are included in this article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The source data behind the graphs in the paper can be found in Supplementary Data 1.
References
Prat, J. New insights into ovarian cancer pathology. Ann. Oncol. 23, x111–x117 (2012).
Matulonis, U. A. et al. Ovarian cancer. Nat. Rev. Dis. Primers 2, 16061 (2016).
Luvero, D., Milani, A. & Ledermann, J. A. Treatment options in recurrent ovarian cancer: latest evidence and clinical potential. Ther. Adv. Med. Oncol. 6, 229–239 (2014).
Helm, C. W. & States, J. C. Enhancing the efficacy of cisplatin in ovarian cancer treatment—could arsenic have a role. J. Ovarian Res. 2, 2 (2009).
Elhelu, M. A. The role of macrophages in immunology. J. Natl. Med. Assoc. 75, 314–317 (1983).
Vinogradov, S., Warren, G. & Wei, X. Macrophages associated with tumors as potential targets and therapeutic intermediates. Nanomedicine (Lond) 9, 695–707 (2014).
Mantovani, A., Sozzani, S., Locati, M., Allavena, P. & Sica, A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 23, 549–555 (2002).
Colvin, E. K. Tumor-associated macrophages contribute to tumor progression in ovarian cancer. Front. Oncol. 4, 137 (2014).
Nowak, M. & Klink, M. The role of tumor-associated macrophages in the progression and chemoresistance of ovarian cancer. Cells 9, 1299 (2020).
Guo, W., Li, Y., Pang, W. & Shen, H. Exosomes: a potential therapeutic tool targeting communications between tumor cells and macrophages. Mol. Ther. 28, 1953–1964 (2020).
Lan, J. et al. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 79, 146–158 (2019).
Yu, C. Y. & Kuo, H. C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 26, 29 (2019).
Li, Y. et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 25, 981–984 (2015).
Chen, W. et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 475, 119–128 (2020).
Zong, Z. H., Du, Y. P., Guan, X., Chen, S. & Zhao, Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J. Exp. Clin. Cancer Res. 38, 437 (2019).
Guan, X. et al. circPUM1 promotes tumorigenesis and progression of ovarian cancer by sponging miR-615-5p and miR-6753-5p. Mol. Ther. Nucleic Acids 18, 882–892 (2019).
Chen, S. et al. Tumor-associated macrophages promote cholangiocarcinoma progression via exosomal Circ_0020256. Cell Death Dis. 13, 94 (2022).
Yu, P. et al. CircTMCO3 promotes gastric cancer progression by regulating miR-577/RAB14 Axis. Cancer Manag. Res. 13, 6079–6088 (2021).
Wang, F., Xu, X., Zhang, N. & Chen, Z. Identification and integrated analysis of hepatocellular carcinoma-related circular RNA signature. Ann. Transl. Med. 8, 294 (2020).
Zhong, Y. et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer 17, 79 (2018).
Zhang, X. et al. MiR-515-5p acts as a tumor suppressor via targeting TRIP13 in prostate cancer. Int. J. Biol. Macromol. 129, 227–232 (2019).
Hamidi, H. & Ivaska, J. Every step of the way: integrins in cancer progression and metastasis. Nat. Rev. Cancer 18, 533–548 (2018).
Ryu, J. et al. Highly expressed integrin-alpha8 induces epithelial to mesenchymal transition-like features in multiple myeloma with early relapse. Mol. Cells 39, 898–908 (2016).
Vo, J. N. et al. The landscape of circular RNA in cancer. Cell 176, 869–881.e813 (2019).
Zheng, Q. et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 7, 11215 (2016).
Yoshioka, Y. et al. Comparative marker analysis of extracellular vesicles in different human cancer types. J. Extracell Vesicles 2, 20424 (2013).
Aichen, Z., Kun, W., Xiaochun, S. & Lingling, T. LncRNA FGD5-AS1 promotes the malignant phenotypes of ovarian cancer cells via targeting miR-142-5p. Apoptosis 26, 348–360 (2021).
Onder, T. T. et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 68, 3645–3654 (2008).
Liu, L., Ye, Y. & Zhu, X. MMP-9 secreted by tumor associated macrophages promoted gastric cancer metastasis through a PI3K/AKT/Snail pathway. Biomed. Pharmacother. 117, 109096 (2019).
Miyoshi, A. et al. Snail and SIP1 increase cancer invasion by upregulating MMP family in hepatocellular carcinoma cells. Br. J. Cancer 90, 1265–1273 (2004).
Brucks, J. A. Ovarian cancer. The most lethal gynecologic malignancy. Nurs. Clin. North Am. 27, 835–845 (1992).
Torre, L. A. et al. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 68, 284–296 (2018).
Dilley, J. et al. Ovarian cancer symptoms, routes to diagnosis and survival—population cohort study in the ‘no screen’ arm of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS). Gynecol. Oncol. 158, 316–322 (2020).
Maia, J., Caja, S., Strano Moraes, M. C., Couto, N. & Costa-Silva, B. Exosome-based cell-cell communication in the tumor microenvironment. Front Cell Dev. Biol. 6, 18 (2018).
Azmi, A. S., Bao, B. & Sarkar, F. H. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 32, 623–642 (2013).
Wu, J. et al. M2 macrophage-derived exosomes facilitate HCC metastasis by transferring alphaM beta2 integrin to tumor cells. Hepatology 73, 1365–1380 (2021).
Yang, Y. et al. M2 macrophage-derived exosomes promote angiogenesis and growth of pancreatic ductal adenocarcinoma by targeting E2F2. Mol. Ther. 29, 1226–1238 (2021).
Huda, M. N. et al. Potential use of exosomes as diagnostic biomarkers and in targeted drug delivery: progress in clinical and preclinical applications. ACS Biomater. Sci. Eng. 7, 2106–2149 (2021).
Zang, X. et al. Exosome-transmitted lncRNA UFC1 promotes non-small-cell lung cancer progression by EZH2-mediated epigenetic silencing of PTEN expression. Cell Death Dis. 11, 215 (2020).
He, A. T., Liu, J., Li, F. & Yang, B. B. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduct Target Ther. 6, 185 (2021).
Ma, R., Ye, X., Cheng, H., Cui, H. & Chang, X. Tumor-derived exosomal circRNA051239 promotes proliferation and migration of epithelial ovarian cancer. Am. J. Transl. Res. 13, 1125–1139 (2021).
Foruzandeh, Z. et al. CircRNAs as potent biomarkers in ovarian cancer: a systematic scoping review. Cell. Mol. Biol. Lett. 26, 41 (2021).
Wang, L. et al. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int. J. Mol. Sci. 20, 5758 (2019).
Wen, L. J., Wang, Y. S. & Tan, P. Y. miR-515-5p inhibits the proliferation, migration and invasion of human breast cancer cells by targeting CBX4. Exp. Ther. Med. 22, 1328 (2021).
Pardo, O. E. et al. miR-515-5p controls cancer cell migration through MARK4 regulation. EMBO Rep. 17, 570–584 (2016).
Desgrosellier, J. S. & Cheresh, D. A. Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer 10, 9–22 (2010).
Gong, Y. Z. et al. Diagnostic and prognostic values of integrin alpha subfamily mRNA expression in colon adenocarcinoma. Oncol. Rep. 42, 923–936 (2019).
Acknowledgements
This work was supported by Hunan Provincial Natural Science Foundation of China (No.2022JJ70106).
Author information
Authors and Affiliations
Contributions
X.M.R.: conceptualization, writing-original draft, methodology, and formal analysis; J.Y.: supervision and validation; Z.Y.W.: data curation and resources; L.Z.X.: investigation and software; Y.P.D.: visualization; K.Q.Z.: funding acquisition, project administration, writing-review and editing. All authors have read and approved the final version of this manuscript to be published.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Written informed consent was required from patients. The Ethics Committee of Hunan Cancer Hospital, The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University approved our study. Animal procedures were approved by the Animal Care and Use Committee of Hunan Cancer Hospital, The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University.
Peer review
Peer review information
Communications Biology thanks Runxi Fu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Gloryn Chia and George Inglis.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ran, XM., Yang, J., Wang, ZY. et al. M2 macrophage-derived exosomal circTMCO3 acts through miR-515-5p and ITGA8 to enhance malignancy in ovarian cancer. Commun Biol 7, 583 (2024). https://doi.org/10.1038/s42003-024-06095-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-06095-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.