Abstract
Tubo-ovarian high-grade serous carcinoma is believed to originate in the fallopian tubes, arising from precursor lesions like serous tubal intraepithelial carcinoma (STIC) and serous tubal intraepithelial lesion (STIL). Adequate diagnosis of these precursors is important, but can be challenging for pathologists. Here we present a deep-learning algorithm that could assist pathologists in detecting STIC/STIL. A dataset of STIC/STIL (n = 323) and controls (n = 359) was collected and split into three groups; training (n = 169), internal test set (n = 327), and external test set (n = 186). A reference standard was set for the training and internal test sets, by a panel review amongst 15 gynecologic pathologists. The training set was used to train and validate a deep-learning algorithm (U-Net with resnet50 backbone) to differentiate STIC/STIL from benign tubal epithelium. The model’s performance was evaluated on the internal and external test sets by ROC curve analysis, achieving an AUROC of 0.98 (95% CI: 0.96–0.99) on the internal test set, and 0.95 (95% CI: 0.90–0.99) on the external test set. Visual inspection of all cases confirmed the accurate detection of STIC/STIL in relation to the morphology, immunohistochemistry, and the reference standard. This model’s output can aid pathologists in screening for STIC, and can contribute towards a more reliable and reproducible diagnosis.
Similar content being viewed by others
Introduction
Tubo-ovarian high-grade serous carcinoma (HGSC) is the most common form of epithelial ovarian cancer (EOC)1,2. It is thought that HGSC originates in the fallopian tube, where different precursor lesions of HGSC have been identified, all harboring TP53 mutations3,4. The clinically most relevant of these is serous tubal intraepithelial carcinoma (STIC), which shows cytomorphological alterations comparable to HGSC, in combination with a characteristic staining pattern in immunohistochemistry (IHC), with aberrant p53 expression and an increased Ki-67 proliferation index3,5,6. Lesions which resemble STIC but fail to meet all diagnostic criteria, are referred to as serous tubal intraepithelial lesions (STIL).
With the discovery that HGSC originates in the fallopian tube, new preventive strategies to decrease EOC risk are being explored. For example, women undergoing abdominal surgery for benign indications can choose to also have their fallopian tubes removed, even if there is no increased EOC risk (opportunistic salpingectomy)7. Women who are at an increased risk for developing HGSC, e.g., BRCA1/2 PV (pathogenic variant) carriers, are counseled on risk-reducing salpingo-oophorectomy (RRSO), or in study settings, on risk-reducing salpingectomy (RRS) with delayed oophorectomy. There are currently multiple prospective trials investigating the safety of this alternative RRS approach, such as the SOROCK (NCT04251053), PROTECTOR (ISRCTN25173360), and the TUBA-WISPII (NCT04294927) studies8,9. A recent study showed that isolated STIC in RRSO specimens is associated with a significantly increased risk of developing peritoneal carcinomatosis and in the study setting of RRS, diagnosis of a STIC is an indication to perform an oophorectomy without delay8,9,10. Though further research is needed to better determine what clinical implications regarding additional diagnostics or treatment is indicated after a diagnosis of STIC, it will depend on a reliable and reproducible diagnosis of these lesions, underlining the importance of an unequivocal diagnosis. The clinical significance of STIL, and its exact role in the pathogenesis of HGSC remains to be elucidated. Currently, STIL has no clinical implications.
STIC can be a challenging diagnosis for pathologists. With an incidence of 0.1% in the average risk population and 3% in the high-risk population, STIC is a rare and therefore unfamiliar diagnosis for many pathologists5,11. Screening for these lesions comes with a considerable workload, as these lesions are often small. The use of the widely adopted sectioning and extensively examining the fimbriated end (SEE-FIM) grossing protocol, which dictates that the entire fallopian tube should be embedded for microscopic examination, helps to maximize the accessible tissue, yet results in numerous hematoxylin and eosin (H&E) stained slides, which the pathologist has to carefully examine12. Another challenge in STIC diagnosis is its moderate reproducibility amongst pathologists, with kappa scores ranging between 0.33 and 0.396,13. Several clinical algorithms have been proposed to aid the pathologists, using various morphological criteria and IHC6,14,15. These algorithms have been a valuable addition and have led to an improvement in reproducibility6. Still, in a recent Delphi study it was found that opinions regarding relevant findings for a diagnosis of STIC vary largely, even among subspecialized gynecologic pathologists5.
With the recent advancements in digital pathology, new possibilities to support pathologists in their diagnosis are emerging. Machine learning approaches, mostly based on deep learning (DL), have been shown to increase accuracy, reproducibility, and efficiency in histopathologic assessment, and have proven their benefit for both screening and classification in various pathology related tasks16. Therefore, we hypothesize that a DL algorithm can assist pathologists with diagnosing STIC. In this study we developed such an algorithm, capable of fully automated analysis of digitalized H&E-stained slides of fallopian tube specimens to detect regions of aberrant epithelium.
Results
Reference standard
The resulting labels from the expert panel are presented in supplementary figure 2. A total of 571 images were reviewed. A label was assigned when three out of five pathologists agreed on the diagnosis. There were 282 images labeled as ‘STIC’ or ‘suspicious for STIC’. 14 images did not receive a final classification as there was no majority vote. The remaining samples were labeled as ‘STIL’ (n = 42), ‘HGSC’ (n = 20), ‘p53 signature’ (n = 17) and ‘normal’ (n = 196). Kappa values between individual participants ranged between 0.35 and 0.64, with a median of 0.53 (Table 1), corresponding to a moderate level of agreement17. When grouping all aberrant entities together, i.e., only making a distinction between normal epithelium and aberrant epithelium, the kappa values ranged between 0.87 and 0.97, with a median of 0.86, corresponding to a strong level of agreement17.
Model evaluation
The model to detect aberrant epithelium reached an AUROC of 0.98 (95% CI, 0.96–0.99) on the internal test set, and an AUROC of 0.95 (95% CI, 0.90–0.99) on the external test set. The ROC curves for both test sets are presented in Fig. 1. In order to transform the model’s segmentation output into class predictions, a threshold needed to be determined along this ROC curve, for when the model predicts STIC/STIL. When opting for the threshold with the highest F1-score, this threshold would be at 0.35 in the internal test set. At this threshold, the model would reach a sensitivity of 0.98 and a specificity of 0.93 and has, at a slide level, three false negatives and thirteen false positives in a set of 327 slides. The external test set, consisting of 186 slides, would then have a sensitivity of 0.98 and a specificity of 0.67, with two false negatives and 37 false positives. However, if we were to select a threshold favoring sensitivity at 0.30, this would result in a sensitivity of 1 and a specificity of 0.82, with 0 false negatives and 36 false positives in the internal test set. For the external test set, the sensitivity would be 0.98 and the specificity 0.55, with one false negative and 60 false positives. A table with incremental threshold to illustrate the effect on the number of false positives, false negatives, precision rate, recall rate, and F1 score is presented in Fig. 1.
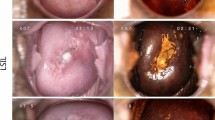
The algorithms segmentation output can be visualized as an overlay image, projected on top of the H&E image. This allows visual inspection of the detected objects, so that the pathologists can assess based on what areas STIC/STIL is predicted. Absence of color thereby means that the model does not predict STIC/STIL. Green indicates that the model predicts STIC/STIL, with a relatively low level of certainty, and red indicates that the model has a high level of certainty that STIC/STIL is present. An overall good alignment was observed between the morphologically aberrant areas, IHC, and the model’s predictions (Fig. 2). The cases resulting in false negative predictions under the 0.35 threshold are shown in Fig. 3. Upon visual inspection, the algorithm did pick up on these lesions, however the probability did not reach the required threshold. In the internal test set, the false negatives consisted of two cases of STIL, and one case of STIC with concomitant HGSC, where the model did detect the HGSC component. The external test set contained two false negatives, that did not reach the 0.35 threshold. One of these cases had been a point of discussion during the case review, whereby there was doubt whether the specimen contained reactive epithelium or STIC. The presence of concomitant HGSC, eventually led the reviewers to classify the lesion as STIC. The HGSC component in this case was again detected. False positives were, amongst other things, observed in areas with increased numbers of lymphocytes, detachments of surface epithelium, or artifacts. A selection of examples is presented in Fig. 4.
Cases A–C are from the internal test set. A1- C1: regions of aberrant epithelium, classified as STIC. A2- C2: model’s prediction, using a color-coded probability map. The absence of color means that the model does not predict STIC/STIL. Green indicates a low certainty in STIC/STIL prediction, and red indicates a high certainty in STIC/STIL prediction. A3-C3: accompanying p53 immunohistochemical stain. A4–C4: accompanying Ki-67 immunohistochemical stain. Cases D–G are from the external test set. D1-G1: regions of aberrant epithelium, classified as STIC. D2-G2: model’s prediction.
Cases A–C are from the internal test set. A1–B1: regions of aberrant epithelium, classified as STIL. A3: regions of aberrant epithelium, classified as STIC with concomitant HGSC. A2- C2: model’s prediction, using a color coded probability map. Green indicates a low certainty in STIC/STIL prediction, and red indicates a high certainty in STIC/STIL prediction. A3-C3: accompanying p53 IHC. A4-C4: accompanying Ki-67 IHC. Cases D–E images from the external test set. D1: region of aberrant epithelium, classified as STIC. D2: model’s prediction. D3: accompanying P53 IHC. D4: accompanying Ki67 IHC. E1: region of aberrant epithelium, classified as STIC. E2: model’s prediction. E3: concomitant HGSC from the same slide. E4: model’s prediction (HGSC adequately detected)
A–C are from the internal test set. A1: region of benign epithelium. B1: epithelium with reactive changes in the background of inflammation. C1: region with artifact (mechanically damaged benign epithelium). A2–C2: model’s prediction, using a color coded probability map. Green indicates a low certainty in STIC/STIL prediction, and red indicates a high certainty in STIC/STIL prediction. D–E are from the external test set. D1: region of benign epithelium. E1: partially destained region (artifact) of benign epithelium. D2–E2: model’s prediction.
Discussion
This study introduces a deep learning (DL) model designed to automatically detect regions of aberrant epithelium (STIC/STIL) in digitalized H&E-stained whole slide images of fallopian tube specimens. To our knowledge, this is the first DL model developed for this application. Through comprehensive testing on two independent test sets, we demonstrate the robust performance and discriminative ability of our algorithm in distinguishing between slides with aberrant epithelium and control slides, achieving high sensitivity and specificity.
The uniqueness of this study lies in its pioneering attempt to improve STIC diagnostics through the application of DL. For this, we developed a U-Net model. U-Net is a widely used and proven method for segmentation tasks in computational pathology, and can be considered the current baseline method. This study stands out as a broad-based study, establishing a large network of international collaboration involving 12 centers from four countries. Despite the rarity of STIC lesions, our study successfully assembled a sizable dataset. To ensure a strong reference standard, we established an international expert group comprising 15 experienced gynecologic pathologists from participating centers who assessed the samples. The resulting DL model exhibited a robust and high-performance outcome, with an AUROC of 0.98 (95% CI, 0.96–0.99) for the internal test set and 0.95 (95% CI, 0.90–0.99) for the external test set.
The limitations of this study are largely associated with the rarity and moderate reproducibility of STIC/STIL. Though the dataset is large, considering the rarity of this lesion, it remains modest in size for developing a DL algorithm and specifically, the number of isolated STIL is low. The input data for training and testing were collected from eight different data sources, encompassing multiple laboratories, staining protocols, and slide scanners. Even with such variability in scanners and staining protocols, our algorithm demonstrated consistent and robust performance on the test sets. Nevertheless, further validation studies on additional datasets, encompassing various staining and sampling methods, will be needed to confirm the generalizability of the model, before this model can be applied in clinical practice. Another challenge is found in the reproducibility of STIC, whereby we know that the diagnosis is prone to interobserver variability6,13. Our expert panel exhibited a strong level of agreement when distinguishing between aberrant and normal epithelium (median k of 0.86), yet agreement levels decreased when differentiating between normal epithelium, high-grade serous carcinoma (HGSC), and various precursor lesions (median k of 0.53), illustrating that the exact boundaries between the various precursor lesions and early HGSC remains challenging even for gynecologic pathologists. Our aim was for this DL model to detect all precursor lesions and not miss any potential STIC. Because the interobserver variability in differentiating STIL and STIC among pathologists is moderate, we chose to include STIL in this detection task. This model, in its current form is thus a detection model and not a classification model. It is not intended to replace the pathologist, but is an additional tool to support the pathologist in this challenging diagnostic task.
The sensitivity and specificity of our model are dependent upon the selected operating threshold. While aiming for the highest F1-score, the optimal operating point was found to be at 0.35. However, in the envisioned setting where pathologists screen for a rare entity, it may be preferable to prioritize high sensitivity (to avoid missing relevant lesions) at the expense of reduced specificity, leading to more false positives that require visual inspection by pathologists. We could thus change the threshold to make the model more sensitive. At a threshold of 0.30, the two missed STIL lesions under the first threshold in the internal test set would be detected, albeit with an additional 23 false positives (out of 327 slides).
Accurate diagnosis of STIC is critical because of the increased risk of peritoneal carcinomatosis associated with isolated STIC lesions. To ensure optimal patient care, it is important that women considering risk-reducing surgeries, either RRSO or RRS with delayed oophorectomy, have access to accurate histologic assessments. Nevertheless, there are currently numerous unresolved issues surrounding STIC. The diagnosis remains challenging and prone to interobserver variability6,13. Crucial research questions regarding the precise role of STIC in the etiology of HGSC and peritoneal carcinomatosis, as well as the potential identification of subtypes within STIC, based on molecular or immunohistochemical profiles, remain unanswered. Finally, there is a lack of sufficient data to guide treatment recommendations, such as the role of follow-up, staging surgery, and chemotherapy in patient management. To address these issues, the development of a reliable and reproducible method for the diagnosis of STIC is of utmost importance.
Deep learning holds immense potential in addressing the diagnostic challenges for STIC. As the digitalization of pathology continues to evolve, DL algorithms can be seamlessly integrated into digital workflows, offering valuable additional screening aids16,18. This workflow augmentation can benefit the analysis of fallopian tube specimens, providing a promising way for STIC detection. Even when adopting a high sensitivity operating point, thereby accepting a more moderate specificity, DL may significantly reduce the time pathologists spend on negative samples. Our study’s DL model exhibited robust performance on two independent test sets, encompassing samples from multiple institutions, suggesting its compatibility with varying institutional setups. However, large-scale multicenter validation is essential to confirm its performance across diverse settings. Furthermore, the model’s efficacy may be further improved through the collection of additional datasets and ongoing technical refinements. While our study primarily focused on detecting aberrant epithelium, we envision that with continued international collaboration and a better understanding of the boundaries between various precursor lesions and early invasive HGSC, DL could well play a role in lesion classification.
In conclusion, our study presents an innovative deep learning model capable of automatically detecting regions of STIC/STIL in digitalized H&E-stained whole slide images of fallopian tubes. This DL model illustrates the potential for deep-learning models, to assist pathologists in reliably identifying regions of aberrant epithelium, enabling them to prioritize their attention and time to the most important samples.
Methods
Materials
A retrospective dataset was collected, comprised of cases of STIC/STIL (n = 323) and benign controls (n = 359). This dataset was split into three groups: a training set, an internal test set and an external test set. The training set (118 STIC/STIL; 51 controls) and internal test set(131 STIC/STIL; 196 controls) were collected from, Radboudumc, the Dutch Nationwide Network of Histopathology and Cytopathology database (PALGA); Kathleen Cuningham Foundation Consortium for research into Familial Breast cancer / Royal Women’s Hospital Melbourne; University of Pennsylvania; Department of Defense (DoD) and Specialized Programs of Research Excellence (SPORE) Ovarian Cancer Omics Consortium; the University of Toronto; the Canisius Wilhelmina Hospital Nijmegen (CWZ), and Rijnstate Hospital Arnhem. Immunohistochemical stains for p53 and Ki-67 were included when available (n = 180). Pathologists may use these stains to assist in differentiating between STIL and STIC. Isolated STIC/STIL, or STIC/STIL with concomitant HGSC from 2011 to 2020 were included. A single slide per patient was selected. Controls consisted of salpingectomy specimens, removed for benign indications, in the age range between 30 and 70. Attention was given to include BRCA1/2 pv carriers amongst the controls, though BRCA status could not be ascertained for all patients. The training set was designated for training and validation, whereby 118 slides (70%) were used for training the algorithm and 51 slides (30%) were used for validation. No samples from the internal test set were seen by the algorithm during the training or validation phases. The external test set (74 cases; 112 controls), consisted of cases from the Johns Hopkins University and controls from the Eurofins PAMM laboratory. Samples from neither of these institutions had previously been seen by the algorithm. An overview of the dataset is provided in Table 2.
Reference standard
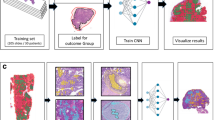
A reference standard was established for the images in the training set and internal test set. Regions of interest from all cases (n = 249), previously diagnosed as (potential) STIC/STIL and a random sample from the controls (n = 247) were selected, based on morphology and immunohistochemistry. A total of 571 still images of these regions were made at 20 times magnification (pixel size 0.5um/pixel). The images were presented to a panel of fifteen experienced gynecologic pathologists, from four different countries and 12 different institutions, using an online pathology viewing platform (grand-challenge.org). The pathologists were randomly split into six groups, consisting of two or three pathologists per group. Each image was reviewed by two groups, with overlap between the groups. Thus, each image was reviewed by five pathologists (supplementary figure 1). Participants were blinded to the original diagnosis. They reviewed the H&E image and, when available, corresponding IHC stains. The pathologists were asked to assign a label to each image, classifying them as either ‘normal/reactive’, ‘STIC’, ‘STIL’, ‘p53 signature’, ‘HGSC’, ‘Suspicious for STIC’, or ‘other’. Though the term ‘suspicious for STIC’ is discouraged for use in diagnostic practice, the term was included in this study setting, to provide an additional option, in case pathologists felt uncomfortable diagnosing STIC, solely based on H&E, when IHC was not available.
The reference standard for the external test set consisted of the original diagnosis at the time of clinical review, which was confirmed by a pathologist and a pathology resident at Radboudumc (MSi and JBo). No discrepancies in this set were identified.
Deep-learning model development
Annotations
Annotations were made in the digitalized whole slide images, using the in-house developed open-source software ASAP (https://github.com/computationalpathologygroup/ASAP). Regions of STIC and STIL were annotated exhaustively, in accordance with the labels that resulted from the reference standard. Other regions, such as invasive cancer, normal epithelium, cystic epithelium, and non-epithelial tissue were annotated sparsely. No hand drawn annotations were made in the control slides.
Deep-learning model development
The development of the deep-learning algorithm consisted of two phases. Phase one focused on the segmentation of all epithelium; differentiating this from all non-epithelial tissue. For this, a U-Net model with a mobilenet-v2 backbone was trained19,20,21. The mobilenet-v2 backbone is a lightweight backbone, which suffices for the task of epithelium segmentation. In phase two, we enhanced the course annotations of aberrant epithelial regions using the epithelial segmentation results from phase one. This process created clear demarcations between these areas and background tissue. We subsequently trained another U-Net segmentation model to differentiate the lesions of interest (regions of aberrant epithelium, classified as STIC or STIL) from normal epithelium, HGSC, and non-epithelial regions. To handle this more complex task, we replaced the Mobilenet-v2 backbone with the larger ResNet50 backbone19,20,22. A graphic representation of the entire training pipeline is shown in Fig. 5.
Training parameters
The input for both U-Net models was a RGB patch of 512 × 512px, with a pixel spacing of 1.0 um/pixel. Patches were randomly sampled from the annotated regions. Care was taken to ensure a balanced set of training patches containing aberrant epithelium and healthy tissue, meaning that half of the mini batch contained aberrant epithelium and the other half healthy tissue. During training, random flipping, rotation, elastic deformation, blurring, brightness (random gamma), color, and contrast changes augmentations were used, in order to improve generalizability. The learning rate was initially set to 1e-4 and multiplied by a factor of 0.5 after every 25 epochs if no increase in performance was observed on the validation set. The networks were initialized with pre-trained weights, trained on ImageNet data. The networks were trained for a maximum of 150 epochs, with 500 iterations per epoch. The mini-batch size was set to 10 per batch, resulting in the network seeing 5000 patches per epoch. Training of the networks was stopped when no improvement of the validation loss was found for 50 epochs. The output of all networks is in the form of C likelihood maps. Pytorch 1.9 in Python 3.8 was used for the development of the algorithm.
Manual hard-negative mining (HNM) was performed during training of both the model from phase one and phase two. At the end of each training session, the model was applied to the full training set at whole-slide level. All regions that were incorrectly segmented were manually annotated as difficult regions for the appropriate class. During the next training session these regions were sampled twice as often, in comparison to the less challenging regions, to allow the model to focus on these difficult regions. While training, we monitored the performance of the model on a validation set using the DICE coefficient. We experimentally noted that after five HNM iterations, no further increase in segmentation performance was observed on the validation set.
Model evaluation
Output of phase one, segmenting all epithelium, was inspected visually, without additional quantitative evaluation. Output of phase two was evaluated by assessing the slide level predictions. To obtain a slide level prediction, we first identified connected components of segmented regions, with a STIC/STIL probability. The average probability of such regions was assigned as the object probability. The highest object probability was thereafter used as the slide level prediction. We subsequently created a receiver operating characteristics curve (ROC-curve) and calculated the corresponding area under the receiver operating characteristics curve (AUROC), using sklearn (scikit-learn.org) in python 3.8. To obtain a confidence interval we performed bootstrapping with 1000 iterations, using Numpy 1.21 and Scipy 1.7.1 in python 3.8. Finally, slides were checked visually to compare the objects that the model detected as aberrant regions, with corresponding morphology and IHC.
Statistics
Kappa values for the reference standard were calculated using IBM SPSS statistics version 27.
Data availability
Images are subject to various data transfer agreements. These images can be requested at the respective pathology institutions. Source codes to train and assess the deep learning model and data from the reference standard are available from the corresponding author on reasonable request. The deep-learning model will be made freely accessible for research purposes and can be accessed on-line (grand-challenge.org), after this manuscript is accepted for publication.
References
Seidman, J. D. et al. The histologic type and stage distribution of ovarian carcinomas of surface epithelial origin. Int. J. Gynecol. Pathol. 23, 41–44 (2004).
Peres, L. C. et al. Invasive epithelial ovarian cancer survival by histotype and disease stage. J. Natl Cancer Inst. 111, 60–68 (2019).
Kuhn, E. et al. Shortened telomeres in serous tubal intraepithelial carcinoma: an early event in ovarian high-grade serous carcinogenesis. Am. J. Surg. Pathol. 34, 829–836 (2010).
Shih, I. M., Wang, Y. & Wang, T. L. The origin of ovarian cancer species and precancerous landscape. Am. J. Pathol. 191, 26–39 (2021).
Bogaerts, J. M. A. et al. Consensus based recommendations for the diagnosis of serous tubal intraepithelial carcinoma: an international Delphi study. Histopathology, (2023).
Visvanathan, K. et al. Diagnosis of serous tubal intraepithelial carcinoma based on morphologic and immunohistochemical features: a reproducibility study. Am. J. Surg. Pathol. 35, 1766–1775 (2011).
Fillon, M. Opportunistic salpingectomy may reduce ovarian cancer risk. CA Cancer J. Clin. 72, 97–99 (2022).
Huh, W. K. et al. NRG-CC008: A nonrandomized prospective clinical trial comparing the non-inferiority of salpingectomy to salpingo-oophorectomy to reduce the risk of ovarian cancer among BRCA1 carriers [SOROCk]. J. Clin. Oncol. 40, TPS10615–TPS10615 (2022).
Steenbeek, M. P. et al. TUBectomy with delayed oophorectomy as an alternative to risk-reducing salpingo-oophorectomy in high-risk women to assess the safety of prevention: the TUBA-WISP II study protocol. Int. J. Gynecol. Cancer, (2023).
Steenbeek, M. P. et al. Risk of peritoneal carcinomatosis after risk-reducing salpingo-oophorectomy: a systematic review and individual patient data meta-analysis. J. Clin. Oncol. 40, 1879–1891 (2022).
Samimi, G., Trabert, B., Geczik, A. M., Duggan, M. A. & Sherman, M. E. Population frequency of serous tubal intraepithelial carcinoma (STIC) in clinical practice using SEE-fim protocol. JNCI Cancer Spectr. 2, pky061 (2018).
Medeiros, F. et al. The tubal fimbria is a preferred site for early adenocarcinoma in women with familial ovarian cancer syndrome. Am. J. Surg. Pathol 30, 230–236 (2006).
Carlson, J. W. et al. Serous tubal intraepithelial carcinoma: diagnostic reproducibility and its implications. Int. J. Gynecol. Pathol. 29, 310–314 (2010).
Meserve, E. E. K., Brouwer, J. & Crum, C. P. Serous tubal intraepithelial neoplasia: the concept and its application. Mod. Pathol. 30, 710–721 (2017).
Perrone, M. E. et al. An alternate diagnostic algorithm for the diagnosis of intraepithelial fallopian tube lesions. Int. J. Gynecol. Pathol. 39, 261–269 (2020).
van der Laak, J., Litjens, G. & Ciompi, F. Deep learning in histopathology: the path to the clinic. Nat. Med. 27, 775–784 (2021).
Altman, D. G. Practical Statistics for Medical Research (1st ed.). (Chapman and Hall/CRC., 1990).
Echle, A. et al. Deep learning in cancer pathology: a new generation of clinical biomarkers. Br. J. Cancer 124, 686–696 (2021).
Ronneberger, O., Fischer, P. & Brox, T. in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. (eds Navab, N., Hornegger, J., Wells, W. M. & Frangi, A. F.) 234–241 (Springer International Publishing).
Segmentation Models Pytorch (GitHub, GitHub repository, 2019).
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A. & Chen, L. C. in 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. 4510–4520.
He, K., Zhang, X., Ren, S. & Sun, J. in 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 770–778.
Acknowledgements
We would like to thank the Kathleen Cuningham Foundation Consortium for research into Familial Breast cancer (kConFab), the Department of Defense (DoD) and Specialized Programs of Research Excellence (SPORE) Ovarian Cancer Omics Consortium, and the Dutch Nationwide Network of Histopathology and Cytopathology database (PALGA), for their contributions to case collection. This study was funded by the Dutch Cancer Society (KWF), grant number 12950. The funder played no role in the study design, data collection, analysis, and interpretation of data, or the writing of this manuscript.
Author information
Authors and Affiliations
Contributions
Conception and design: J.Bo., J.M.B., M.Si., J.H., J.L. Collection and assembly of data: J.Bo., M.Si., J.H., J.L., M.Bo., M.St., J.Ba., Y.W.C., M.D., T.N., L.S., I.M.S., T.R.S., G.T., R.V., M.V. Data analysis and interpretation: All authors. Manuscript writing: All authors. Final approval of manuscript: All authors. Accountable for all aspects of the work: All authors.
Corresponding author
Ethics declarations
Competing interests
JL was a member of the advisory boards of Philips, the Netherlands and ContextVision, Sweden, and received research funding from Philips, the Netherlands, ContextVision, Sweden, and Sectra, Sweden in the last five years. He is chief scientific officer (CSO) and shareholder of Aiosyn BV, the Netherlands.
Ethics
This project was reviewed and approved by the research ethics committee at the Radboud University Medical Center (Radboudumc) (#: 2019-5879). The need for informed patient consent was waived by the ethics committee.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bogaerts, J.M.A., Bokhorst, JM., Simons, M. et al. Deep learning detects premalignant lesions in the Fallopian tube. npj Womens Health 2, 11 (2024). https://doi.org/10.1038/s44294-024-00016-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44294-024-00016-0