Featured
-
-
Article
| Open Access Structure-guided discovery of anti-CRISPR and anti-phage defense proteins
Structure-guided discovery of anti-CRISPR and anti-phage defense proteins
Bacteria use various defense systems to protect themselves from phage infection, and phages have evolved diverse counter-defense measures to overcome host defenses. Here, the authors use protein structural similarity and gene co-occurrence analyses for identification of new anti-phage and counter-defense systems.
- Ning Duan
- , Emily Hand
- & Akintunde Emiola
-
Article
| Open Access Local energetic frustration conservation in protein families and superfamilies
Local energetic frustration conservation in protein families and superfamilies
Energetic local frustration in proteins may have been positively selected by evolution when related to function such as ligand binding, allostery and other. Here the authors present a methodology to analyze local frustration patterns within protein families and superfamilies.
- Maria I. Freiberger
- , Victoria Ruiz-Serra
- & Alfonso Valencia
-
Article
| Open Access Resurrecting ancestral antibiotics: unveiling the origins of modern lipid II targeting glycopeptides
Resurrecting ancestral antibiotics: unveiling the origins of modern lipid II targeting glycopeptides
Glycopeptide antibiotics (GPAs) are microbial natural products synthesized by multiple enzymes, including a nonribosomal peptide synthetase for assembly of the peptide core. Here, the authors use computational techniques to infer a gene set for biosynthesis of an ancestral GPA, produce the peptide in a microbial host, and provide insights into the evolution of key enzymatic domains.
- Mathias H. Hansen
- , Martina Adamek
- & Nadine Ziemert
-
Article
| Open Access Ongoing shuffling of protein fragments diversifies core viral functions linked to interactions with bacterial hosts
Ongoing shuffling of protein fragments diversifies core viral functions linked to interactions with bacterial hosts
Proteins are composed of distinct functional domains, each serving a specific role. Here, Smug et al. show that phages are able to shuffle fragments of their proteins and this predominantly occurs in proteins involved in bacterial host interactions.
- Bogna J. Smug
- , Krzysztof Szczepaniak
- & Rafał J. Mostowy
-
Article
| Open Access Functional annotation of enzyme-encoding genes using deep learning with transformer layers
Functional annotation of enzyme-encoding genes using deep learning with transformer layers
Functional annotation of open reading frames in microbial genomes remains substantially incomplete. Here, Kim et al. present a deep learning model that utilizes transformer layers as a neural network architecture to predict specific catalytic functions for enzyme-encoding genes of unknown function.
- Gi Bae Kim
- , Ji Yeon Kim
- & Sang Yup Lee
-
Article
| Open Access Dynamic characterization and interpretation for protein-RNA interactions across diverse cellular conditions using HDRNet
Dynamic characterization and interpretation for protein-RNA interactions across diverse cellular conditions using HDRNet
Predicting dynamic RNA-RBP interactions in diverse cell lines is an important challenge in unravelling RNA function and post-transcriptional regulatory mechanisms. Here, authors develop HDRNet, an end-to-end deep-learning-based framework for accurately predicting dynamic RBP binding events across various cellular conditions.
- Haoran Zhu
- , Yuning Yang
- & Xiangtao Li
-
Article
| Open Access Machine learning coarse-grained potentials of protein thermodynamics
Machine learning coarse-grained potentials of protein thermodynamics
Understanding protein dynamics is a complex scientific challenge. Here, authors construct coarse-grained molecular potentials using artificial neural networks, significantly accelerating protein dynamics simulations while preserving their thermodynamics.
- Maciej Majewski
- , Adrià Pérez
- & Gianni De Fabritiis
-
Article
| Open Access Deep transfer learning for inter-chain contact predictions of transmembrane protein complexes
Deep transfer learning for inter-chain contact predictions of transmembrane protein complexes
Membrane proteins are encoded by approximately a quarter of human genes. Here, the authors propose a deep transfer learning method for predicting inter-chain residue-residue contacts of transmembrane protein complexes.
- Peicong Lin
- , Yumeng Yan
- & Sheng-You Huang
-
Article
| Open Access Discovering functionally important sites in proteins
Discovering functionally important sites in proteins
An important step in understanding and using proteins is to identify the residues that are important for function. The authors present a machine-learning based method to predict functional sites that leverages and combines the information available in protein sequences and structures.
- Matteo Cagiada
- , Sandro Bottaro
- & Kresten Lindorff-Larsen
-
Article
| Open Access Predicting the antigenic evolution of SARS-COV-2 with deep learning
Predicting the antigenic evolution of SARS-COV-2 with deep learning
SARS-CoV-2’s rapid evolution threatens public health. Here, authors present a deep learning approach to forecast high-risk mutations that may appear in the future, aiding vaccine development and enhancing preparedness against future variants.
- Wenkai Han
- , Ningning Chen
- & Xin Gao
-
Article
| Open Access Machine learning optimization of candidate antibody yields highly diverse sub-nanomolar affinity antibody libraries
Machine learning optimization of candidate antibody yields highly diverse sub-nanomolar affinity antibody libraries
Therapeutic antibody discovery is time and cost-intensive. Here, the authors develop a machine learning-driven method enabling accelerated design of large and diverse single-chain variable fragments with high binding efficiency, especially at high levels of diversity.
- Lin Li
- , Esther Gupta
- & Matthew E. Walsh
-
Article
| Open Access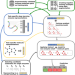 A general model to predict small molecule substrates of enzymes based on machine and deep learning
A general model to predict small molecule substrates of enzymes based on machine and deep learning
For many enzymes, it is unknown which primary and/or secondary reactions they catalyze. Here, the authors use machine and deep learning to develop a general model for the prediction of enzyme-small molecule substrate pairs and make the resulting model available through a webserver.
- Alexander Kroll
- , Sahasra Ranjan
- & Martin J. Lercher
-
Article
| Open Access The RESP AI model accelerates the identification of tight-binding antibodies
The RESP AI model accelerates the identification of tight-binding antibodies
High-affinity antibodies are often identified through directed evolution but deep leaning methods hold great promise. Here the authors report RESP, a pipeline for efficient identification of high affinity antibodies, and apply this to the PD-L1 antibody Atezolizumab.
- Jonathan Parkinson
- , Ryan Hard
- & Wei Wang
-
Article
| Open Access Dynamic spatiotemporal determinants modulate GPCR:G protein coupling selectivity and promiscuity
Dynamic spatiotemporal determinants modulate GPCR:G protein coupling selectivity and promiscuity
G protein coupled receptors (GPCRs) can couple to different Gα protein subfamilies either selectively or promiscuously. Here, the authors use computational approach to show that selectivity determinants are at the periphery of the GPCR—G protein interface and that promiscuous GPCRs more frequently sample the common rather than selective contacts.
- Manbir Sandhu
- , Aaron Cho
- & Nagarajan Vaidehi
-
Article
| Open Access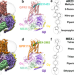 Activation and signaling mechanism revealed by GPR119-Gs complex structures
Activation and signaling mechanism revealed by GPR119-Gs complex structures
Agonists selectively targeting GPR119 hold promise for treating metabolic disorders. Here, authors reveal that GPR119 adopts a non-canonical consensus structural scaffold with an extended ligand-binding pocket for chemically different agonists.
- Yuxia Qian
- , Jiening Wang
- & Anna Qiao
-
Article
| Open Access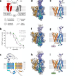 Structural basis of organic cation transporter-3 inhibition
Structural basis of organic cation transporter-3 inhibition
The current work reports the structure of the human organic cation transporter 3 (OCT3 / SLC22A3) and provides the structural basis of its inhibition by two specific inhibitors, decynium-22 and corticosterone.
- Basavraj Khanppnavar
- , Julian Maier
- & Harald H. Sitte
-
Article
| Open Access Deciphering microbial gene function using natural language processing
Deciphering microbial gene function using natural language processing
The function of many microbial genes is yet unknown. Here the authors repurposed natural language processing algorithms to explore “gene semantics” and infer function for thousands of genes with defense and secretion systems found to have the most discovery potential.
- Danielle Miller
- , Adi Stern
- & David Burstein
-
Article
| Open Access KSTAR: An algorithm to predict patient-specific kinase activities from phosphoproteomic data
KSTAR: An algorithm to predict patient-specific kinase activities from phosphoproteomic data
Kinases are important drug targets, but predicting their activities from phosphoproteomics data remains challenging. While many existing prediction tools rely on phosphosite-specific quantitative data, Crowl et al. develop a kinase activity prediction algorithm that requires no phosphosite quantification.
- Sam Crowl
- , Ben T. Jordan
- & Kristen M. Naegle
-
Article
| Open Access Computational identification of HCV neutralizing antibodies with a common HCDR3 disulfide bond motif in the antibody repertoires of infected individuals
Computational identification of HCV neutralizing antibodies with a common HCDR3 disulfide bond motif in the antibody repertoires of infected individuals
Identifying determinants of broadly neutralizing antibodies against hepatitis C virus (HCV) may guide HCV vaccine design. Here, the authors discover new anti-HCV antibodies using computational screening and analyze the amino acid composition and sequence-structure relationships in this antibody family.
- Nina G. Bozhanova
- , Andrew I. Flyak
- & Jens Meiler
-
Article
| Open Access Helical structure motifs made searchable for functional peptide design
Helical structure motifs made searchable for functional peptide design
Here, we present TP-DB; a pattern-based search engine based on 1.67 million helices from the Protein Database (PDB). We demonstrate the utility of TP-DB in identifying microbe-specific antigens, as well as the design of antimicrobial peptides and Protein-protein interaction blockers.
- Cheng-Yu Tsai
- , Emmanuel Oluwatobi Salawu
- & Lee-Wei Yang
-
Article
| Open Access Co-evolution based machine-learning for predicting functional interactions between human genes
Co-evolution based machine-learning for predicting functional interactions between human genes
With the rise in number of eukaryotic species being fully sequenced, large scale phylogenetic profiling can give insights on gene function, Here, the authors describe a machine-learning approach that integrates co-evolution across eukaryotic clades to predict gene function and functional interactions among human genes.
- Doron Stupp
- , Elad Sharon
- & Yuval Tabach
-
Article
| Open Access Epistatic Net allows the sparse spectral regularization of deep neural networks for inferring fitness functions
Epistatic Net allows the sparse spectral regularization of deep neural networks for inferring fitness functions
Finding a biologically-relevant inductive bias for training DNNs on large fitness landscapes is challenging. Here, the authors propose a method called Epistatic Net that improves DNN prediction accuracy and interpretation speed by integrating the knowledge that higher-order epistatic interactions are usually sparse.
- Amirali Aghazadeh
- , Hunter Nisonoff
- & Kannan Ramchandran
-
Article
| Open Access Activation pathway of a G protein-coupled receptor uncovers conformational intermediates as targets for allosteric drug design
Activation pathway of a G protein-coupled receptor uncovers conformational intermediates as targets for allosteric drug design
G protein-coupled receptors (GPCRs) are a critical target in modern drug development across a wide range of indications. Here the authors provide a comprehensive characterization of a typical GPCR, the angiotensin II (AngII) type 1 receptor (AT1R), and provide insight into its activation mechanism that suggest avenues for the design of allosteric GPCR modulators.
- Shaoyong Lu
- , Xinheng He
- & Jian Zhang
-
Article
| Open Access flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions
flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions
The authors present flDPnn, a computational tool for disorder and disorder function predictions from protein sequences. flDPnn was assessed with the data from the “Critical Assessment of Protein Intrinsic Disorder Prediction” experiment and on an independent and low-similarity test dataset, which show that flDPnn offers accurate predictions of disorder, fully disordered proteins and four common disorder functions.
- Gang Hu
- , Akila Katuwawala
- & Lukasz Kurgan
-
Article
| Open Access Structure-based protein function prediction using graph convolutional networks
Structure-based protein function prediction using graph convolutional networks
The rapid increase in the number of proteins in sequence databases and the diversity of their functions challenge computational approaches for automated function prediction. Here, the authors introduce DeepFRI, a Graph Convolutional Network for predicting protein functions by leveraging sequence features extracted from a protein language model and protein structures.
- Vladimir Gligorijević
- , P. Douglas Renfrew
- & Richard Bonneau
-
Article
| Open Access Protein design and variant prediction using autoregressive generative models
Protein design and variant prediction using autoregressive generative models
The ability to design functional sequences is central to protein engineering and biotherapeutics. Here the authors introduce a deep generative alignment-free model for sequence design applied to highly variable regions and design and test a diverse nanobody library with improved properties for selection experiments.
- Jung-Eun Shin
- , Adam J. Riesselman
- & Debora S. Marks
-
Article
| Open Access Large-scale discovery of protein interactions at residue resolution using co-evolution calculated from genomic sequences
Large-scale discovery of protein interactions at residue resolution using co-evolution calculated from genomic sequences
Our understanding of the residue-level details of protein interactions remains incomplete. Here, the authors show sequence coevolution can be used to infer interacting proteins with residue-level details, including predicting 467 interactions de novo in the Escherichia coli cell envelope proteome.
- Anna G. Green
- , Hadeer Elhabashy
- & Debora S. Marks
-
Article
| Open Access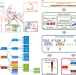 Inferring the molecular and phenotypic impact of amino acid variants with MutPred2
Inferring the molecular and phenotypic impact of amino acid variants with MutPred2
Identifying variants capable of causing genetic disease is challenging. The authors use semisupervised learning to predict pathogenic missense variants and their impacts on protein structure and function, enabling a molecular mechanism-driven approach to studying different types of human disease.
- Vikas Pejaver
- , Jorge Urresti
- & Predrag Radivojac
-
Article
| Open Access Multisecond ligand dissociation dynamics from atomistic simulations
Multisecond ligand dissociation dynamics from atomistic simulations
Protein-ligand unbinding processes are out of reach for atomistic simulations due to time-scale involved. Here the authors demonstrate an approach relying on dissipation-corrected targeted molecular dynamics that enables to provide binding and unbinding rates with a speed-up of several orders of magnitude.
- Steffen Wolf
- , Benjamin Lickert
- & Gerhard Stock
-
Article
| Open Access Environmental conditions shape the nature of a minimal bacterial genome
Environmental conditions shape the nature of a minimal bacterial genome
Minimal bacterial genomes still contain hundreds of genes of unknown function. Here the authors use in silico annotation methods and identify the environmental factors shaping a minimal genome.
- Magdalena Antczak
- , Martin Michaelis
- & Mark N. Wass
-
Article
| Open Access Improving the diagnostic yield of exome- sequencing by predicting gene–phenotype associations using large-scale gene expression analysis
Improving the diagnostic yield of exome- sequencing by predicting gene–phenotype associations using large-scale gene expression analysis
A genetic diagnosis remains unattainable for many individuals with a rare disease because of incomplete knowledge about the genetic basis of many diseases. Here, the authors present the web-based tool GADO (GeneNetwork Assisted Diagnostic Optimization) that uses public RNA-seq data for prioritization of candidate genes.
- Patrick Deelen
- , Sipko van Dam
- & Lude Franke
-
Article
| Open Access Domain insertion permissibility-guided engineering of allostery in ion channels
Domain insertion permissibility-guided engineering of allostery in ion channels
Allostery is a fundamental principle of protein regulation that remains challenging to engineer. Here authors screen human Inward Rectifier K + Channel Kir2.1 for permissibility to domain insertions and propose that differential permissibility is a metric of latent allosteric capacity in Kir2.1.
- Willow Coyote-Maestas
- , Yungui He
- & Daniel Schmidt
-
Article
| Open Access A recurrent point mutation in PRKCA is a hallmark of chordoid gliomas
A recurrent point mutation in PRKCA is a hallmark of chordoid gliomas
Chordoid glioma is a slow growing diencephalic tumor whose mutational landscape is poorly characterized. Here, the authors perform whole-exome and RNA-sequencing and find that 15 of 16 chordoid glioma cases studied harbor the same PRKCA mutation which results in enhanced proliferation.
- Shai Rosenberg
- , Iva Simeonova
- & Marc Sanson
-
Article
| Open Access Multi-omics analysis reveals neoantigen-independent immune cell infiltration in copy-number driven cancers
Multi-omics analysis reveals neoantigen-independent immune cell infiltration in copy-number driven cancers
Neoantigen load has been associated with tumour immune infiltration. Here, the authors show that while this is true for tumours with recurrent mutations, cancers with recurrent CNAs show neoantigen-independent infiltration driven by cytokine production downstream of the DNA damage sensor ATM.
- Daniel J. McGrail
- , Lorenzo Federico
- & Nidhi Sahni
-
Article
| Open Access Discovery of a proteolytic flagellin family in diverse bacterial phyla that assembles enzymatically active flagella
Discovery of a proteolytic flagellin family in diverse bacterial phyla that assembles enzymatically active flagella
So far no enzymatic activity has been attributed to flagellin, the major component of bacterial flagella. Here the authors use bioinformatic analysis and identify a metallopeptidase insertion in flagellins from 74 bacterial species and show that recombinant flagellin and flagellar filaments have proteolytic activity.
- Ulrich Eckhard
- , Hina Bandukwala
- & Andrew C. Doxey
-
Article
| Open Access An integrated bioinformatics platform for investigating the human E3 ubiquitin ligase-substrate interaction network
An integrated bioinformatics platform for investigating the human E3 ubiquitin ligase-substrate interaction network
Protein stability modulation by E3 ubiquitin ligases is an important layer of functional regulation, but screening for E3 ligase-substrate interactions is time-consuming and costly. Here, the authors take an in silico naïve Bayesian classifier approach to integrate multiple lines of evidence for E3-substrate prediction, enabling prediction of the proteome-wide human E3 ligase interaction network.
- Yang Li
- , Ping Xie
- & Fuchu He
-
Article
| Open Access Exchange pathways of plastoquinone and plastoquinol in the photosystem II complex
Exchange pathways of plastoquinone and plastoquinol in the photosystem II complex
Plastoquinone (PLQ) shuttles electrons between photosystem II (PSII) and cytochrome b6f. Here the authors perform molecular dynamics simulations and propose that PLQ enters the exchange cavity of PSII by a promiscuous diffusion mechanism whereby three different channels each act as entry and exit points.
- Floris J. Van Eerden
- , Manuel N. Melo
- & Siewert J. Marrink
-
Article
| Open Access Extreme multifunctional proteins identified from a human protein interaction network
Extreme multifunctional proteins identified from a human protein interaction network
Proteins are sometimes implicated in separate and seemingly unrelated processes, so called moonlighting functions. Here the authors use bioinformatics tools to identify extreme multifunctional proteins and define a signature of extreme multifunctionality.
- Charles E. Chapple
- , Benoit Robisson
- & Christine Brun

 One substrate many enzymes virtual screening uncovers missing genes of carnitine biosynthesis in human and mouse
One substrate many enzymes virtual screening uncovers missing genes of carnitine biosynthesis in human and mouse