Featured
-
-
Article
| Open Access Role of UPF1-LIN28A interaction during early differentiation of pluripotent stem cells
Role of UPF1-LIN28A interaction during early differentiation of pluripotent stem cells
UPF1 and LIN28A are RNA-binding proteins involved in post-transcriptional regulation and cell differentiation. Here, authors report that they interact with each other via specific domains and regulate ectodermal specialization of human pluripotent stem cells.
- Seungwon Jung
- , Seung Hwan Ko
- & Jungwook Hwang
-
Article
| Open Access Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex
Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex
Here the authors show how the MTREC core protein Red1 binds to and sequesters Pla1 from the 3’-end processing machinery to hyperadenylate cryptic unstable transcripts and target them to the exosome for efficient degradation.
- Komal Soni
- , Anusree Sivadas
- & Tamás Fischer
-
Article
| Open Access Temporal-iCLIP captures co-transcriptional RNA-protein interactions
Temporal-iCLIP captures co-transcriptional RNA-protein interactions
Dynamic RNA-protein interactions govern the co-transcriptional packaging of RNA polymerase II derived transcripts. Here the authors use temporal-iCLIP which combines transcriptional synchronisation with UV cross-linking of RNA-protein complexes to reveal dynamic RNA-protein interactions during the early phases of transcription and beyond.
- Ross A. Cordiner
- , Yuhui Dou
- & Torben Heick Jensen
-
Article
| Open Access The PNUTS-PP1 complex acts as an intrinsic barrier to herpesvirus KSHV gene expression and replication
The PNUTS-PP1 complex acts as an intrinsic barrier to herpesvirus KSHV gene expression and replication
The PNUTS-PP1 complex directly binds to RNA, and interacts with polymerase II and RNA processing factors to control transcriptional elongation rates and slow polymerase II after polyadenylation sites to promote termination. Using a genome-wide CRISPR screen, Devlin et al. identify this complex as a critical suppressor of herpesvirus KSHV gene expression. They further provide evidence that PNUTS-PP1 controls elongation both downstream and upstream of polyadenylation sites on specific viral genes.
- Anne M. Devlin
- , Ashutosh Shukla
- & Nicholas K. Conrad
-
Article
| Open Access Altered tRNA processing is linked to a distinct and unusual La protein in Tetrahymena thermophila
Altered tRNA processing is linked to a distinct and unusual La protein in Tetrahymena thermophila
La proteins are conserved factors critical for the maturation of RNA polymerase III transcripts. In the ciliate T. thermophila and related alveolates, La proteins have a novel domain arrangement and are linked to a distinct pre-tRNA processing pathway.
- Kyra Kerkhofs
- , Jyoti Garg
- & Mark A. Bayfield
-
Article
| Open Access Gene-specific nonsense-mediated mRNA decay targeting for cystic fibrosis therapy
Gene-specific nonsense-mediated mRNA decay targeting for cystic fibrosis therapy
The W1282X nonsense mutation in the CFTR gene causes cystic fibrosis by reducing its mRNA and functional protein levels. Here the authors developed antisense-oligonucleotide cocktails that restore CFTR protein function by gene-specific stabilization of CFTR mRNA.
- Young Jin Kim
- , Tomoki Nomakuchi
- & Adrian R. Krainer
-
Article
| Open Access The methyl phosphate capping enzyme Bmc1/Bin3 is a stable component of the fission yeast telomerase holoenzyme
The methyl phosphate capping enzyme Bmc1/Bin3 is a stable component of the fission yeast telomerase holoenzyme
There exists a remarkable evolutionary breadth of telomerase holoenzyme components among eukaryotes. Here the authors report that the fission yeast methyl phosphate capping enzyme Bin3/Bmc1 regulates telomerase activity by promoting holoenzyme formation.
- Jennifer Porat
- , Moaine El Baidouri
- & Mark A. Bayfield
-
Article
| Open Access Cellular variability of nonsense-mediated mRNA decay
Cellular variability of nonsense-mediated mRNA decay
Here the author developed a single-cell reporter system to identify cell-to-cell variability of nonsense-mediated mRNA decay (NMD). This approach provides a sensitive tool to investigate cellular heterogeneity of NMD in various physiological conditions.
- Hanae Sato
- & Robert H. Singer
-
Article
| Open Access Terminal uridyltransferase 7 regulates TLR4-triggered inflammation by controlling Regnase-1 mRNA uridylation and degradation
Terminal uridyltransferase 7 regulates TLR4-triggered inflammation by controlling Regnase-1 mRNA uridylation and degradation
Terminal uridyltransferase 7 (TUT7) adds U-tails on diverse RNAs to promote degradation. Here the authors show that TUT7 is induced upon LPS treatment in macrophages and promotes decay of Regnase-1, thereby regulating the expression of a subset of cytokines, including IL-6.
- Chia-Ching Lin
- , Yi-Ru Shen
- & Li-Chung Hsu
-
Article
| Open Access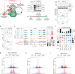 SMG5-SMG7 authorize nonsense-mediated mRNA decay by enabling SMG6 endonucleolytic activity
SMG5-SMG7 authorize nonsense-mediated mRNA decay by enabling SMG6 endonucleolytic activity
Degradation of nonsense mediated mRNA decay (NMD) substrates is carried out by two seemingly independent pathways, SMG6-mediated endonucleolytic cleavage and/or SMG5-SMG7-induced accelerated deadenylation. Here the authors show that SMG5-SMG7 maintain NMD activity by permitting SMG6 activation.
- Volker Boehm
- , Sabrina Kueckelmann
- & Niels H. Gehring
-
Article
| Open Access TSSC4 is a component of U5 snRNP that promotes tri-snRNP formation
TSSC4 is a component of U5 snRNP that promotes tri-snRNP formation
The correct assembly and recycling of the multicomponent spliceosome remains largely elusive. Here, the authors show that a previously uncharacterized protein TSSC4 associates with de novo formed spliceosomal U5 snRNP as well as with a post-splicing U5-PRPF19 particle, and that TSSC4 is important for assembly of the splicing competent tri-snRNP.
- Klára Klimešová
- , Jitka Vojáčková
- & David Staněk
-
Article
| Open Access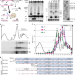 No-Go Decay mRNA cleavage in the ribosome exit tunnel produces 5′-OH ends phosphorylated by Trl1
No-Go Decay mRNA cleavage in the ribosome exit tunnel produces 5′-OH ends phosphorylated by Trl1
In the No-Go decay mRNA surveillance pathway, mRNAs containing stalled ribosomes are cleaved by an endoribonuclease. Here the authors show the endonucleolytic cleavage on the artificial No-Go decay target mRNAs, revealing downstream decay process by Trl1 kinase and the 5′ to 3′ exonuclease Dxo1.
- Albertas Navickas
- , Sébastien Chamois
- & Lionel Benard
-
Article
| Open Access Oxidation and alkylation stresses activate ribosome-quality control
Oxidation and alkylation stresses activate ribosome-quality control
The mRNA-surveillance pathway of no-go decay (NGD) is a eukaryotic ribosome-based-quality-control process that targets transcripts that stall the ribosome. Here the authors show no-go decay (NGD) and ribosome-quality control (RQC) pathways are activated by mRNAs damaged by alkylation and oxidation stress.
- Liewei L. Yan
- , Carrie L. Simms
- & Hani S. Zaher
-
Article
| Open Access RST1 and RIPR connect the cytosolic RNA exosome to the Ski complex in Arabidopsis
RST1 and RIPR connect the cytosolic RNA exosome to the Ski complex in Arabidopsis
Cytosolic RNA degradation by the RNA exosome requires the Ski complex. Here the authors show that the proteins RST1 and RIPR assist the RNA exosome and the Ski complex in RNA degradation, thereby preventing the production of secondary siRNAs from endogenous mRNAs.
- Heike Lange
- , Simon Y. A. Ndecky
- & Dominique Gagliardi
-
Article
| Open Access Molecular interactions between Hel2 and RNA supporting ribosome-associated quality control
Molecular interactions between Hel2 and RNA supporting ribosome-associated quality control
Ribosome-associated quality control (RQC) pathways monitor and respond to stalling of the translating ribosome. Here the authors show that the ribosome associated RQC factor Hel2/ZNF598, an E3 ubiquitin ligase, generally interacts with mRNAs in the vicinity of stop codons.
- Marie-Luise Winz
- , Lauri Peil
- & David Tollervey
-
Article
| Open Access The H/ACA complex disrupts triplex in hTR precursor to permit processing by RRP6 and PARN
The H/ACA complex disrupts triplex in hTR precursor to permit processing by RRP6 and PARN
Telomerase RNA (hTR) is transcribed as a 3′-extended precursor. Here the authors examine the processing of hTR precursors of various lengths and show that processing occurs in distinct steps involving different nucleases PARN and RRP6.
- Chi-Kang Tseng
- , Hui-Fang Wang
- & Peter Baumann
-
Article
| Open Access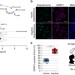 Dedicated surveillance mechanism controls G-quadruplex forming non-coding RNAs in human mitochondria
Dedicated surveillance mechanism controls G-quadruplex forming non-coding RNAs in human mitochondria
G-rich RNAs encoded in mitochondrial DNA are prone to form four-stranded structures called G-quadruplexes (G4s). Here the authors show using in vitro and in vivo approaches that GRSF1 promotes melting of G4 RNA structures in mtRNAs, thus leading to their decay by the hSuv3–PNPase complex.
- Zbigniew Pietras
- , Magdalena A. Wojcik
- & Roman J. Szczesny
-
Article
| Open Access Tailing and degradation of Argonaute-bound small RNAs protect the genome from uncontrolled RNAi
Tailing and degradation of Argonaute-bound small RNAs protect the genome from uncontrolled RNAi
While RNA interference is a highly conserved mechanism of gene regulation, how Argonaute-bound small RNAs are targeted for degradation is not well understood. Here the authors show that Cid14 and Cid16 target Argonaute-bound small RNAs for degradation and protect the genome from uncontrolled RNAi activity.
- Paola Pisacane
- & Mario Halic
-
Article
| Open Access ATP hydrolysis by UPF1 is required for efficient translation termination at premature stop codons
ATP hydrolysis by UPF1 is required for efficient translation termination at premature stop codons
Nonsense-mediated mRNA decay (NMD) is a quality control pathway that recognizes and degrades transcripts harbouring nonsense mutations. Here the authors show that the ATPase activity of UPF1 mediates functional interactions between the NMD machinery and ribosomes required for efficient ribosome release at premature termination codons.
- Lucas D. Serdar
- , DaJuan L. Whiteside
- & Kristian E. Baker
-
Article
| Open Access Structural insights into ribosomal rescue by Dom34 and Hbs1 at near-atomic resolution
Structural insights into ribosomal rescue by Dom34 and Hbs1 at near-atomic resolution
mRNA surveillance is essential to maintain homeostasis in eukaryotes and is activated by mRNAs lacking a stop codon. Here the authors describe a high resolution cryo-EM structure of a nonstop complex that shows how arrested ribosome recognition is achieved during Dom34-mediated mRNA surveillance.
- Tarek Hilal
- , Hiroshi Yamamoto
- & Christian M.T. Spahn
-
Article
| Open Access Interrogating the degradation pathways of unstable mRNAs with XRN1-resistant sequences
Interrogating the degradation pathways of unstable mRNAs with XRN1-resistant sequences
Degradation of messenger RNA is a key regulatory step in controlling eukaryotic gene expression. Here the authors present xrFrag, a molecular tool to interrogate the extent and directionality of mRNA turnover by the detection of stabilized decay intermediates produced by several common decay pathways.
- Volker Boehm
- , Jennifer V. Gerbracht
- & Niels H. Gehring
-
Article
| Open Access Hyperphosphorylation amplifies UPF1 activity to resolve stalls in nonsense-mediated mRNA decay
Hyperphosphorylation amplifies UPF1 activity to resolve stalls in nonsense-mediated mRNA decay
Gene expression is regulated by a range of mechanisms, including post-translational modifications such as phosphorylation. Here the authors present evidence for a feedback mechanism whereby hyperphosphorylation of UPF1 in response to delays in nonsense-mediated decay enhances recruitment of mRNA decay machinery.
- Sébastien Durand
- , Tobias M. Franks
- & Jens Lykke-Andersen
-
Article |
 Quality control of spliced mRNAs requires the shuttling SR proteins Gbp2 and Hrb1
Quality control of spliced mRNAs requires the shuttling SR proteins Gbp2 and Hrb1
In eukaryotic cells, export of unprocessed pre-mRNAs is prevented by the nuclear surveillance machinery. Here Hackmann et al.identify the SR proteins Gbp2 and Hrb1 as two novel quality control factors for spliced mRNAs that determine their degradation or nuclear export.
- Alexandra Hackmann
- , Haijia Wu
- & Heike Krebber

 Exon-junction complex association with stalled ribosomes and slow translation-independent disassembly
Exon-junction complex association with stalled ribosomes and slow translation-independent disassembly