Abstract
The Sparsely Annotated Region and Organ Segmentation (SAROS) dataset was created using data from The Cancer Imaging Archive (TCIA) to provide a large open-access CT dataset with high-quality annotations of body landmarks. In-house segmentation models were employed to generate annotation proposals on randomly selected cases from TCIA. The dataset includes 13 semantic body region labels (abdominal/thoracic cavity, bones, brain, breast implant, mediastinum, muscle, parotid/submandibular/thyroid glands, pericardium, spinal cord, subcutaneous tissue) and six body part labels (left/right arm/leg, head, torso). Case selection was based on the DICOM series description, gender, and imaging protocol, resulting in 882 patients (438 female) for a total of 900 CTs. Manual review and correction of proposals were conducted in a continuous quality control cycle. Only every fifth axial slice was annotated, yielding 20150 annotated slices from 28 data collections. For the reproducibility on downstream tasks, five cross-validation folds and a test set were pre-defined. The SAROS dataset serves as an open-access resource for training and evaluating novel segmentation models, covering various scanner vendors and diseases.
Similar content being viewed by others
Background & Summary
Medical imaging plays a crucial role in the diagnosis and treatment of various diseases. Computed tomography (CT) imaging is one of the most commonly used imaging modalities, allowing for detailed visualization of internal organs and structures. In recent years, the use of CT imaging for body composition analysis (BCA) has gained increasing attention in clinical research. However, developing accurate and efficient segmentation algorithms for CT images remains a challenging task, mainly due to the limited availability of publicly accessible datasets with high-quality annotations.
Many published works on BCA were conducted on in-house data1,2,3,4 and kept their annotations private or considered only a very specific part of the body5. Commonly used locations for manual body composition assessments in the clinical routine are the C35,6 and L37,8,9,10,11,12 vertebrae, which implies that only single slices in the abdomen and head/neck are measured. Recently published datasets have expanded the pool of data and labels available, including different annotated anatomical landmarks and structures. The TotalSegmentator dataset13,14,15,16 provided segmentations for 117 anatomical structures such as organs, vessels, and specific bones and muscles. CT Volumes with Multiple Organ Segmentations (CT-ORG)17 offered segmentations of some organs and a large-scale segmentation of the bones. AbdomenCT-1K targeted segmentations of four abdominal organs, while the Whole Abdominal Organ Dataset (WORD)18 published segmentations of 16 abdominal organs. Moreover, coding challenges also contributed to the rising number of datasets. The Liver Tumor Segmentation Benchmark (LiTS)19 and the Kidney Tumor Segmentation Challenge (KiTS)20,21 focused on annotations for liver and kidney tumors, respectively. The Lung CT Segmentation Challenge (LCTSC)22,23,24 provided thoracic organs and spinal cord segmentations, while the aim of the Lung Nodule Analysis Challenge 2016 (LUNA16)25,26 was the segmentation of lung lobes and nodules. The Combined Healthy Abdominal Organ Segmentation (CHAOS)27,28 and the Multi-Modality Abdominal Multi-Organ Segmentation Challenge 2022 (AMOS22)29 both provided abdominal organ segmentations for CT and Magnetic Resonance (MR) imaging. The Fast and Low-Resource Semi-supervised Abdominal Organ Segmentation 2022 (FLARE22)30,31 also focused on abdominal organs but used semi-supervised learning for fast and low-resource segmentation. FLARE2332 extended this concept by adding tumor segmentation in these abdominal regions. Additionally, the Head and Neck Autosegmentation challenge33 used the Public Domain Database for Computational Anatomy (PDDCA) segmentations for small head and neck organs and bones.
In most datasets, the focus of the annotation is primarily on organs or pathologies, leaving the rest of the body unexplained. For example, the muscle segmentations from the TotalSegmentator dataset do not cover all the muscle groups of the body. Therefore, the focus of these annotations is different and hardly usable for deriving BCA biomarkers. Furthermore, other structures such as the abdominal cavity, the subcutaneous tissue, and the mediastinum are not publicly available.
In this work, we present the Sparsely Annotated Region and Organ Segmentation (SAROS) dataset using publicly available data from The Cancer Imaging Archive (TCIA)22. The goal of this dataset is to provide a large open-access annotated CT dataset for building automated BCA pipelines for the whole body3,4. In contrast to other openly available datasets, this dataset provides large-scale annotation of body regions, including the subcutaneous tissue, all muscles and bones, the abdominal and thoracic cavities, the mediastinum, and the pericardium. Abdominal, thoracic, and head and neck organs such as liver, lungs, or esophagus were not included, since they are already available in many publicly available imaging datasets. However, the dataset offers annotations of smaller organs such as the thyroid, submandibular and parotid glands. Furthermore, SAROS provides segmentations of the breast implant, which is valuable for improving the differentiation of subcutaneous tissue and could also contribute to more accurate diagnoses of breast pathologies. Additionally, the dataset also includes segmentations of the brain and spinal cord, as well as the segmentation of body parts such as the head, torso, and left/right arms and legs. Another difference with existing datasets is that the SAROS segmentations collectively cover all body voxels. Interested readers can find more information on the applicability of these segmentations for BCA in the Technical Validation section.
The dataset consists of 900 CTs, split into five pre-defined cross-validation folds and a test set, each consisting of 150 CTs. In total, 20,157 slices were annotated with two different label sets. The dataset creation process as well as the relationship to prior work3 is shown in Fig. 1. SAROS was gathered from TCIA data sources, annotation candidates were generated using existing in-house BCA models, manual revision was conducted, and manual and automatic quality control was performed.
Methods
Study population and data conversion
The cohort used in SAROS was built using available imaging data from various collections of TCIA. All available image series on TCIA were searched for keywords in the series description as well as metadata in DICOM by using their REST API. Those keywords were used to restrict the scan region, e.g. lung, thorax, abdomen, or whole-body. In addition, only imaging series with a soft-tissue reconstruction kernel were included. Furthermore, terms for the selection of the contrast agent phases such as arterial or portal-venous were also used. Three groups of imaging series were created based on the captured body region: abdominal, thoracic, and whole-body. Scans from the abdomen and thorax group were mostly contrast-enhanced, whereas scans from the whole-body group were mostly non-contrast-enhanced. For each body region, a total of 300 CTs were collected, resulting in 300 patients with abdominal scans (150 female), 283 patients with thoracic scans (138 female), and 299 patients with whole-body scans (150 female). Ethical approval was not required for this study, as it exclusively utilizes data already available on TCIA. In collaboration with TCIA, a new collection was established, based on previously published data but enriched with new segmentations.
In total, 900 CTs were compiled from 28 TCIA data collections. A detailed overview of how many CTs were drawn from each data collection and for each group is shown in Table 1. Abdomen and thorax scans were easily acquired from 6 and 5 data collections, respectively, while whole-body scans were rarer and thus scattered across 22 data collections. Still, most of the whole-body scans are truncated at the upper legs. As previously mentioned, the CTs were selected randomly across all search results and there was no attempt to reduce the number of required data collections.
The downloaded DICOM data from TCIA was converted to the Neuroimaging Informatics Technology Initiative (NIfTI)34 format using the SimpleITK35 library.
Segmentation
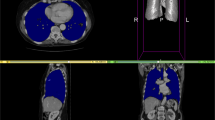
SAROS complements the imaging data with two segmentation files. First, the body-region segmentation represents anatomical structures in the body, e.g. abdominal cavity, muscles, pericardium, and many more. Second, the body-parts segmentation represents a coarse grouping of parts of the body, namely both arms and legs, head, and torso. Exemplary slices from annotated cases can be found in Fig. 2.
Exemplary slices of the annotated CT cases. On the left side, the sparse annotation scheme is visible in the coronal and sagittal slices, since only every fifth axial slice was annotated. On the top, the body regions label set is shown (e.g. red = subcutaneous tissue; green = muscles; blue = thoracic cavity; yellow = abdominal cavity), on the bottom, the corresponding body parts label set is shown (red = torso; yellow = right leg; blue = left leg; green = head; pink = right arm; cyan = left arm).
In previous work, an internal dataset from data in the clinical routine was created to train deep learning segmentation networks to enable fully automated body composition analysis3. This internal dataset initially covered only contrast-enhanced abdominal scans but was extended over time to also support whole-body CTs, contrast-enhanced and non-contrast-enhanced CTs, as well as more semantic regions in order to extract additional biomarkers and to increase the stability of the model predictions3. The internal dataset can be requested from the University Hospital Essen by sending an email with the subject “Request: access to Internal BCA/SAROS dataset” to the corresponding researcher René Hosch (rene.hosch@uk-essen.de) at the Institute for Artificial Intelligence in Medicine (IKIM) with a short summary of the intended use of the dataset. However, as the dataset contains private patient data, it cannot be made publicly available for legal reasons and access can only be granted on specific request. It should also be noted that the internal dataset and the model based on it were only used as a fundament for the initial annotation of the SAROS dataset. Thus, the SAROS dataset provides an independent and larger manually segmented dataset than the internal dataset.
The body-region segmentation in SAROS was annotated with the help of existing models trained on the internal dataset. For this purpose, the image data was resampled to 5 mm slice thickness, as these models are based on 3D network architectures and were trained on 5 mm CTs. In each CT, an annotation for every fifth slice was proposed, which was reviewed and refined by a human reader using ITK Snap36. Every other slice was set to an ignore label (numeric value 255), which resulted in a sparse annotation of the CT scans. This approach drastically reduced the required annotation time compared to starting from scratch, since many proposed contours were already precise enough, and only regions with high uncertainty had to be refined. For example, partial volume effects in 5 mm CTs or abnormal anatomy are a common source of errors for the decision boundaries. In case of strong beam hardening artifacts, mainly in the region of dentition or pelvis due to metallic implants, the slices in question were set completely to an ignore label by the human reviewer. After reviewing and refining all proposed annotations, a quality control team ensured the validity of the annotations and reiterated the refinement process if required. On the one hand, manual control by experienced reviewers was done to identify annotation errors caused by lack of experience or imprecise label definition. On the other hand, an automatic approach was used to identify errors. More details on this are available in the Technical Validation section.
For the body-part segmentation, the refined and reviewed annotations were used to generate a body mask with pre-annotated part labels using simple rules on the body-regions segmentation and a connected component analysis. Suppose that a slice is below the abdominal cavity and has exactly two connected components, it can easily be assumed that those components correspond to the left and right leg. Similarly, a slice showing the abdominal or thoracic cavity with two or three connected components most likely also shows the left and/or right arm. Thus the annotation effort was again greatly reduced, since only a few slices had to be modified where an automatic derivation was not possible.
Split definition
In order to foster reproducibility, five cross-validation folds as well as a test set were predefined, consisting of 150 samples each. Users who want to use a train/validation/test approach without cross-validation, can use “fold-1” for validation and the union of “fold-2”, “fold-3”, “fold-4”, and “fold-5” for training (see Data Records section). Since only 18 CTs in total contain a breast implant label, these were evenly distributed using stratified random sampling. The remaining cases were randomly assigned to the six groups, ensuring that CTs from the same patient were assigned to the same split.
Data Records
The annotated data is stored at TCIA in the collection “SAROS - A large, heterogeneous, and sparsely annotated segmentation dataset on CT imaging data (SAROS)”22,37. The segmentations for each CT are stored in a separate directory with the name “case_xxx” (“xxx” is a zero-padded three-digit case index starting with zero), where the body regions segmentation file “body-regions.nii.gz” and the body parts segmentation file “body-parts.nii.gz” are available. The segmentation files always contain fully annotated axial slices or completely ignored axial slices (numeric value 255). Table 2 contains both label sets (body regions and body parts) including the enumeration index corresponding to the numerical value of the respective label.
A comma-separated value (CSV) file, named “Segmentation Info”, is provided with information about each case, e.g. information about the TCIA identifiers, gender, and CT orientation. Additionally, the column “split” states if the case belongs to one of the five pre-defined cross-validation folds (“fold-1”, …, “fold-5”) or the test set (“test”).
Technical Validation
During the annotation process, a quality control team consisting of senior annotators, a data scientist, and a senior radiologist with 7 years of experience in abdominal imaging, refined and reviewed segmentations first manually and afterward automatically. Manual quality control was mainly based on the experience of the human readers and the internal annotation guidelines. The automatic quality control implemented a few test cases regarding the presence or absence of labels, unusual instance counts for a specific label or neighbor label constraints. For instance, in the abdominal cavity region, the existing models sometimes made tiny mistakes in vessels with high density, due to calcifications or the presence of a contrast agent, and predicted it to be bone. Those errors were sometimes missed by human readers and thus a 2D connected component analysis was performed in order to identify potential instances with invalid neighbors. Additionally, it was ensured that both annotations, “body-parts.nii.gz” and “body-regions.nii.gz”, match pixel-perfect regarding background and foreground labels.
In Table 3, label statistics are shown for each cross-validation fold, the test set, and the overall dataset. In total 900 CTs were annotated, each split contains 150 CTs, and breast implant labels are evenly distributed among all splits.
Baseline models
Two baseline models were trained to provide benchmarkss for the segmentation of body parts and body regions for the SAROS dataset. The nnUNet16 framework was chosen for this purpose, as it is auto-configuring and requires little adjustment. The models were trained using a 2D nnUNet with 5-fold cross validation using the splits outlined in the “Data Records” section. The scripts used for this purpose are available on GitHub (https://github.com/UMEssen/saros-dataset/tree/main/training). The README file in the repository provides guidelines for training and evaluation of the models using the Dice score38 and the Normalized Surface Dice (NSD)39. The NSD calculates the frequency at which the surface distance between volumes measures under 3 mm, a metric previously utilized by TotalSegmentator13. The scores were calculated as the average of the single labels over the CT scans and are presented in Table 4 for both models.
Further discussion of methods
Datasets similar to SAROS have already been successfully used for building a BCA pipeline for multiple research studies3,4,40,41,42,43. However, none of these datasets were released open-source.
An exemplary pipeline for BCA from these studies is presented in Fig. 3. First, the “body-regions” segmentation is used to predict semantic regions. The regions can then be subclassified using Hounsfield Unit (HU) thresholding to distinguish the localization of adipose tissue7,44,45,46 and to refine the muscle segmentation11. Both the muscular and the adipose tissue can be derived based on known and established HU thresholds (adipose tissue −190 to −30 HU, muscular tissue −29 to 150 HU)47, or could be computed by material decomposition algorithms using either dual-energy CTs (DECT) or photon-counting CTs (PCCT)48,49. In research projects, it has been shown that especially ratios between bone, muscular tissue, and different adipose tissue types are important biomarkers for diagnostic and prognostic endpoints40,41,42,43.
Usage Notes
The segmentations and the CSV file provided in the SAROS TCIA collection22,37 are freely accessible and can be downloaded from the TCIA collection page. However, the underlying CT images originate from a combination of datasets, all of which require the user to have a registered TCIA account. Moreover, some datasets require the user to sign a TCIA Data Usage Agreement. A source code repository is available on Github (https://github.com/UMEssen/saros-dataset) with a download.py script to easily download and convert the CT scans to resampled NIfTI images. After running the download.py script, the CT image is downloaded, resampled, and stored as “image.nii.gz”. If the option–save-original-image was provided, the original CT image without resampling is additionally stored as “image-original.nii.gz”.
Code availability
A script to download and resample the data is available on GitHub (https://github.com/UMEssen/saros-dataset).
References
Islam, S. et al. Fully automated deep-learning section-based muscle segmentation from CT images for sarcopenia assessment. Clin. Radiol. 77, e363–e371 (2022).
Zopfs, D. et al. Evaluating body composition by combining quantitative spectral detector computed tomography and deep learning-based image segmentation. Eur. J. Radiol. 130, 109153 (2020).
Koitka, S., Kroll, L., Malamutmann, E., Oezcelik, A. & Nensa, F. Fully automated body composition analysis in routine CT imaging using 3D semantic segmentation convolutional neural networks. Eur. Radiol. 31, 1795–1804 (2021).
Haubold, J. et al. BOA: A CT-Based Body and Organ Analysis for Radiologists at the Point of Care. Invest. Radiol. 59, (2024).
Wahid, K. A. et al. Muscle and adipose tissue segmentations at the third cervical vertebral level in patients with head and neck cancer. Sci. Data 9, 470 (2022).
Zopfs, D. et al. Two-dimensional CT measurements enable assessment of body composition on head and neck CT. Eur. Radiol. 32, 6427–6434 (2022).
Cespedes Feliciano, E. M. et al. Evaluation of automated computed tomography segmentation to assess body composition and mortality associations in cancer patients. J. Cachexia Sarcopenia Muscle 11, 1258–1269 (2020).
Ha, J. et al. Development of a fully automatic deep learning system for L3 selection and body composition assessment on computed tomography. Sci. Rep. 11, 21656 (2021).
Nowak, S. et al. Fully Automated Segmentation of Connective Tissue Compartments for CT-Based Body Composition Analysis: A Deep Learning Approach. Invest. Radiol. 55, 357–366 (2020).
Nowak, S. et al. End-to-end automated body composition analyses with integrated quality control for opportunistic assessment of sarcopenia in CT. Eur. Radiol. 32, 3142–3151 (2022).
Chandarana, H. et al. Association of body composition parameters measured on CT with risk of hospitalization in patients with Covid-19. Eur. J. Radiol. 145, 110031 (2021).
Chandarana, H. et al. Visceral adipose tissue in patients with COVID-19: risk stratification for severity. Abdom. Radiol. N. Y. 46, 818–825 (2021).
Wasserthal, J. et al. TotalSegmentator: Robust Segmentation of 104 Anatomic Structures in CT Images. Radiol. Artif. Intell. 5, e230024 (2023).
Wasserthal, J. Dataset with segmentations of 104 important anatomical structures in 1204 CT images. Zenodo https://doi.org/10.5281/zenodo.6802614 (2022).
Wasserthal, J. Dataset with segmentations of 117 important anatomical structures in 1228 CT images. Zenodo https://doi.org/10.5281/zenodo.10047292 (2023).
Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J. & Maier-Hein, K. H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18, 203–211 (2021).
Rister, B., Yi, D., Shivakumar, K., Nobashi, T. & Rubin, D. L. CT-ORG, a new dataset for multiple organ segmentation in computed tomography. Sci. Data 7, 381 (2020).
Luo, X. et al. WORD: A large scale dataset, benchmark and clinical applicable study for abdominal organ segmentation from CT image. Med. Image Anal. 82, 102642 (2022).
Bilic, P. et al. The Liver Tumor Segmentation Benchmark (LiTS). Med. Image Anal. 84, 102680 (2023).
Heller, N. et al. The state of the art in kidney and kidney tumor segmentation in contrast-enhanced CT imaging: Results of the KiTS19 challenge. Med. Image Anal. 67, 101821 (2021).
Heller, N. et al. C4KC KiTS Challenge Kidney Tumor Segmentation Dataset. The Cancer Imaging Archive https://doi.org/10.7937/TCIA.2019.IX49E8NX (2019).
Clark, K. et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 26, 1045–1057 (2013).
Yang, J. et al. Data from Lung CT Segmentation Challenge 2017 (LCTSC). The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2017.3R3FVZ08 (2017).
Yang, J. et al. Autosegmentation for thoracic radiation treatment planning: A grand challenge at AAPM 2017. Med. Phys. 45, 4568–4581 (2018).
Armato III, S. G. et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A Completed Reference Database of Lung Nodules on CT Scans. Med. Phys. 38, 915–931 (2011).
Tang, H., Zhang, C. & Xie, X. Automatic Pulmonary Lobe Segmentation Using Deep Learning. 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), 1225-1228, https://doi.org/10.1109/ISBI.2019.8759468 (2019).
Kavur, A. E. et al. CHAOS Challenge - combined (CT-MR) healthy abdominal organ segmentation. Med. Image Anal. 69, 101950 (2021).
Kavur, A. E., Selver, M. A., Dicle, O., Barış, M. & Gezer, N. S. CHAOS - Combined (CT-MR) Healthy Abdominal Organ Segmentation Challenge Data. Zenodo https://doi.org/10.5281/zenodo.3431873 (2019).
Ji, Y. et al. AMOS: A Large-Scale Abdominal Multi-Organ Benchmark for Versatile Medical Image Segmentation. Preprint at https://doi.org/10.48550/arXiv.2208.12041 (2022).
Ma, J. MICCAI FLARE22 Challenge Dataset (50 Labeled Abdomen CT Scans). Zenodo https://doi.org/10.5281/zenodo.7860267 (2023).
Ma, J. et al. Unleashing the Strengths of Unlabeled Data in Pan-cancer Abdominal Organ Quantification: the FLARE22 Challenge. Preprint at https://doi.org/10.48550/arXiv.2308.05862 (2023).
MICCAI FLARE23: Fast, Low-resource, and Accurate oRgan and Pan-cancer sEgmentation in Abdomen CT. https://codalab.lisn.upsaclay.fr/competitions/12239 (2023).
Raudaschl, P. F. et al. Evaluation of segmentation methods on head and neck CT: Auto-segmentation challenge 2015. Med. Phys. 44, 2020–2036 (2017).
Li, X., Morgan, P. S., Ashburner, J., Smith, J. & Rorden, C. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J. Neurosci. Methods 264, 47–56 (2016).
Lowekamp, B. C., Chen, D. T., Ibáñez, L. & Blezek, D. The Design of SimpleITK. Front. Neuroinformatics 7, (2013).
Yushkevich, P. A. et al. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. NeuroImage 31, 1116–1128 (2006).
Koitka, S. et al. SAROS - A large, heterogeneous, and sparsely annotated segmentation dataset on CT imaging data (SAROS). The Cancer Imaging Archive https://doi.org/10.25737/SZ96-ZG60 (2023).
Dice, L. R. Measures of the Amount of Ecologic Association Between Species. Ecology 26, 297–302 (1945).
Nikolov, S. et al. Clinically Applicable Segmentation of Head and Neck Anatomy for Radiotherapy: Deep Learning Algorithm Development and Validation Study. J. Med. Internet Res. 23, e26151 (2021).
Kroll, L., Nassenstein, K., Jochims, M., Koitka, S. & Nensa, F. Assessing the Role of Pericardial Fat as a Biomarker Connected to Coronary Calcification-A Deep Learning Based Approach Using Fully Automated Body Composition Analysis. J. Clin. Med. 10, 356 (2021).
Hosch, R. et al. Biomarkers extracted by fully automated body composition analysis from chest CT correlate with SARS-CoV-2 outcome severity. Sci. Rep. 12, 16411 (2022).
Keyl, J. et al. Deep learning-based assessment of body composition and liver tumour burden for survival modelling in advanced colorectal cancer. J. Cachexia Sarcopenia Muscle 14, 545–552 (2023).
Kroll, L. et al. CT-derived body composition analysis could possibly replace DXA and BIA to monitor NET-patients. Sci. Rep. 12, 13419 (2022).
Grainger, A. T. et al. Deep Learning-based Quantification of Abdominal Subcutaneous and Visceral Fat Volume on CT Images. Acad. Radiol. 28, 1481–1487 (2021).
Lee, S. J. et al. Fully automated segmentation and quantification of visceral and subcutaneous fat at abdominal CT: application to a longitudinal adult screening cohort. Br. J. Radiol. 91, 20170968 (2018).
Hwang, J. J. & Pak, K. Development of automated segmentation of visceral adipose tissue in computed tomography. Eur. J. Radiol. 157, 110559 (2022).
Aubrey, J. et al. Measurement of skeletal muscle radiation attenuation and basis of its biological variation. Acta Physiol. 210, 489–497 (2014).
Molwitz, I. et al. Fat Quantification in Dual-Layer Detector Spectral Computed Tomography: Experimental Development and First In-Patient Validation. Invest. Radiol. 57, 463–469 (2022).
Molwitz, I. et al. Skeletal muscle fat quantification by dual-energy computed tomography in comparison with 3T MR imaging. Eur. Radiol. 31, 7529–7539 (2021).
Kostakoglu, L. et al. A Phase II Study of 3′-Deoxy-3′- 18 F-Fluorothymidine PET in the Assessment of Early Response of Breast Cancer to Neoadjuvant Chemotherapy: Results from ACRIN 6688. J. Nucl. Med. 56, 1681–1689 (2015).
Kinahan, P., Muzi, M., Bialecki, B. & Coombs, L. Data from ACRIN-FLT-Breast. The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2017.OL20ZMXG (2017).
Kinahan, P., Muzi, M., Bialecki, B. & Coombs, L. Data from the ACRIN 6685 Trial HNSCC-FDG-PET/CT. TCIA https://doi.org/10.7937/K9/TCIA.2016.JQEJZZNG (2019).
Lowe, V. J. et al. Multicenter Trial of [18 F]fluorodeoxyglucose Positron Emission Tomography/Computed Tomography Staging of Head and Neck Cancer and Negative Predictive Value and Surgical Impact in the N0 Neck: Results From ACRIN 6685. J. Clin. Oncol. 37, 1704–1712 (2019).
Kinahan, P., Muzi, M., Bialecki, B., Herman, B. & Coombs, L. Data from the ACRIN 6668 Trial NSCLC-FDG-PET. The Cancer Imaging Archive https://doi.org/10.7937/TCIA.2019.30ILQFCL (2019).
Machtay, M. et al. Prediction of Survival by [18 F]Fluorodeoxyglucose Positron Emission Tomography in Patients With Locally Advanced Non–Small-Cell Lung Cancer Undergoing Definitive Chemoradiation Therapy: Results of the ACRIN 6668/RTOG 0235 Trial. J. Clin. Oncol. 31, 3823–3830 (2013).
Patnana, M., Patel, S. & Tsao, A. S. Data from Anti-PD-1 Immunotherapy Lung. The Cancer Imaging Archive https://doi.org/10.7937/TCIA.2019.ZJJWB9IP (2019).
Patnana, M., Patel, S. & Tsao, A. Anti-PD-1 Immunotherapy Melanoma Dataset. The Cancer Imaging Archive https://doi.org/10.7937/TCIA.2019.1AE0QTCU (2019).
Saltz et al. Stony Brook University COVID-19 Positive Cases. The Cancer Imaging Archive https://doi.org/10.7937/TCIA.BBAG-2923 (2021).
National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). The Clinical Proteomic Tumor Analysis Consortium Cutaneous Melanoma Collection (CPTAC-CM). The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2018.ODU24GZE (2018).
National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). The Clinical Proteomic Tumor Analysis Consortium Lung Squamous Cell Carcinoma Collection (CPTAC-LSCC). The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2018.6EMUB5L2 (2018).
National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). The Clinical Proteomic Tumor Analysis Consortium Lung Adenocarcinoma Collection (CPTAC-LUAD). The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2018.PAT12TBS (2018).
National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). The Clinical Proteomic Tumor Analysis Consortium Pancreatic Ductal Adenocarcinoma Collection (CPTAC-PDA). The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2018.SC20FO18 (2018).
National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). The Clinical Proteomic Tumor Analysis Consortium Uterine Corpus Endometrial Carcinoma Collection (CPTAC-UCEC). The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2018.3R3JUISW (2019).
Grossberg, A. et al. HNSCC. The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2020.A8SH-7363 (2020).
MICCAI/M.D. Anderson Cancer Center Head and Neck Quantitative Imaging Working Group. Matched computed tomography segmentation and demographic data for oropharyngeal cancer radiomics challenges. Sci. Data 4, 170077 (2017).
Grossberg, A. J. et al. Imaging and clinical data archive for head and neck squamous cell carcinoma patients treated with radiotherapy. Sci. Data 5, 180173 (2018).
Bosch, W. R., Straube, W. L., Matthews, J. W. & Purdy, J. A. Data From Head-Neck_Cetuximab. The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2015.7AKGJUPZ (2015).
Ang, K. K. et al. Randomized Phase III Trial of Concurrent Accelerated Radiation Plus Cisplatin With or Without Cetuximab for Stage III to IV Head and Neck Carcinoma: RTOG 0522. J. Clin. Oncol. 32, 2940–2950 (2014).
G, A. I., Samuel et al. Data From LIDC-IDRI. The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2015.LO9QL9SX (2015).
Armato, S. G. et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A Completed Reference Database of Lung Nodules on CT Scans: The LIDC/IDRI thoracic CT database of lung nodules. Med. Phys. 38, 915–931 (2011).
Li, P. et al. A Large-Scale CT and PET/CT Dataset for Lung Cancer Diagnosis. The Cancer Imaging Archive https://doi.org/10.7937/TCIA.2020.NNC2-0461 (2020).
Napel, S. & Plevritis, S. K. NSCLC Radiogenomics: Initial Stanford Study of 26 Cases. The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2014.X7ONY6B1 (2014).
Bakr, S. et al. Data for NSCLC Radiogenomics Collection. The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2017.7HS46ERV (2017).
Bakr, S. et al. A radiogenomic dataset of non-small cell lung cancer. Sci. Data 5, 180202 (2018).
Gevaert, O. et al. Non–Small Cell Lung Cancer: Identifying Prognostic Imaging Biomarkers by Leveraging Public Gene Expression Microarray Data—Methods and Preliminary Results. Radiology 264, 387–396 (2012).
Aerts, H. J. W. L. et al. Data From NSCLC-Radiomics. The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2015.PF0M9REI (2019).
Aerts, H. J. W. L. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, 4006 (2014).
Aerts, H. J. W. L. et al. Data From NSCLC-Radiomics-Genomics. The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2015.L4FRET6Z (2015).
Roth, H. et al. Data From Pancreas-CT. The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2016.TNB1KQBU (2016).
Roth, H. R. et al. DeepOrgan: Multi-level Deep Convolutional Networks for Automated Pancreas Segmentation. in Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015 (eds. Navab, N., Hornegger, J., Wells, W. M. & Frangi, A.) 556–564 (Springer International Publishing, 2015).
Beichel, R. R. et al. Data From QIN-HEADNECK. The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2015.K0F5CGLI (2015).
Fedorov, A. et al. DICOM for quantitative imaging biomarker development: a standards based approach to sharing clinical data and structured PET/CT analysis results in head and neck cancer research. PeerJ 4, e2057 (2016).
Vallières, M., Freeman, C. R., Skamene, S. R. & El Naqa, I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2015.7GO2GSKS (2015).
Vallières, M., Freeman, C. R., Skamene, S. R. & El Naqa, I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys. Med. Biol. 60, 5471–5496 (2015).
Zuley, M. L. et al. The Cancer Genome Atlas Head-Neck Squamous Cell Carcinoma Collection (TCGA-HNSC). The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2016.LXKQ47MS (2016).
Erickson, B. J. et al. The Cancer Genome Atlas Liver Hepatocellular Carcinoma Collection (TCGA-LIHC). The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2016.IMMQW8UQ (2016).
Albertina, B. et al. The Cancer Genome Atlas Lung Adenocarcinoma Collection (TCGA-LUAD). The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2016.JGNIHEP5 (2016).
Kirk, S. et al. The Cancer Genome Atlas Lung Squamous Cell Carcinoma Collection (TCGA-LUSC). The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2016.TYGKKFMQ (2016).
Lucchesi, F. R. & Aredes, N. D. The Cancer Genome Atlas Stomach Adenocarcinoma Collection (TCGA-STAD). The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2016.GDHL9KIM (2016).
Erickson, B. J., Mutch, D., Lippmann, L. & Jarosz, R. The Cancer Genome Atlas Uterine Corpus Endometrial Carcinoma Collection (TCGA-UCEC). The Cancer Imaging Archive https://doi.org/10.7937/K9/TCIA.2016.GKJ0ZWAC (2016).
Acknowledgements
Special thanks to the Annotation Lab (IKIM, University Hospital Essen, Germany, https://annotationlab.ikim.nrw) who segmented the imaging data. Without any doubt, this project would not have been possible without the annotation effort of: Katharina Abel, Kerem Bakacak, Hajra Batool, Hannah Baumeister, Irina Kallmeyer, Sara Erma Kaya, Aylin Korkmaz, Christian Marc Kramer, Alina Krenzin, Daniel van Landeghem, Pia Nath, Anton Schily, Musa Shah, and Johannes Vogelsang.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
Sven Koitka designed the study, analyzed the data, built the dataset, conducted quality control, and wrote the initial and revised manuscript. René Hosch designed the study, collected the data and revised the manuscript. Lennard Kroll and Natalie van Landeghem annotated the data, supervised the annotation process, conducted quality control, and revised the manuscript. Giulia Baldini finalized the data and revised the manuscript. Olivia B. Pollok reviewed and corrected the annotations and revised the manuscript. Obioma Pelka, Moon S. Kim, Johannes Haubold, Jens Kleesiek, and Felix Nensa supervised the study and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Koitka, S., Baldini, G., Kroll, L. et al. SAROS: A dataset for whole-body region and organ segmentation in CT imaging. Sci Data 11, 483 (2024). https://doi.org/10.1038/s41597-024-03337-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03337-6