Abstract
A depleted antimicrobial drug pipeline combined with an increasing prevalence of Gram-negative ‘superbugs’ has increased interest in nano therapies to treat antibiotic resistance. As cubosomes and polymyxins disrupt the outer membrane of Gram-negative bacteria via different mechanisms, we herein examine the antimicrobial activity of polymyxin-loaded cubosomes and explore an alternative strategy via the polytherapy treatment of pathogens with cubosomes in combination with polymyxin. The polytherapy treatment substantially increases antimicrobial activity compared to polymyxin B-loaded cubosomes or polymyxin and cubosomes alone. Confocal microscopy and neutron reflectometry suggest the superior polytherapy activity is achieved via a two-step process. Firstly, electrostatic interactions between polymyxin and lipid A initially destabilize the outer membrane. Subsequently, an influx of cubosomes results in further membrane disruption via a lipid exchange process. These findings demonstrate that nanoparticle-based polytherapy treatments may potentially serve as improved alternatives to the conventional use of drug-loaded lipid nanoparticles for the treatment of “superbugs”.
Similar content being viewed by others
Introduction
The World Health Organization (WHO) has declared antimicrobial resistance to be among the top ten global public health threats1. Gram-negative bacteria such as Acinetobacter baumannii, Pseudomonas aeruginosa, and Klebsiella pneumoniae are all included in the WHO Priority Pathogens List urgently requiring novel treatment options2. A major reason why the aforementioned organisms are so difficult to treat is that they are surrounded by an outer membrane (OM) that acts as an impermeable barrier, preventing antibiotics from reaching their targets inside the bacteria3,4. For this reason, polymyxins, available clinically as polymyxin B (PMB) and polymyxin E are considered a last-line therapy for the treatment of antimicrobial-resistant Gram-negative organisms5,6. The polymyxins initially bind to lipopolysaccharide (LPS) located within the outer leaflet of OM of Gram-negative bacteria, causing considerable OM disorganization and ultimately cell death7. However, reports of polymyxin resistance globally have increased, threatening the utility of these important antibiotics8,9 and leaving only few new antibiotics currently under development. Therefore, there is an urgent need for novel therapeutic options that may potentially be applicable for use in combating these antimicrobial-resistant Gram-negative “superbugs”.
The use of existing polymyxins10,11 in combination with other antibiotics12,13,14,15,16 or nonantibiotic adjuvants17,18,19,20 to improve bacterial killing is a promising strategy21. Another approach for overcoming antimicrobial resistance involves loading antimicrobials into nanoparticles which transports the antibiotics to their bacterial target sites22,23,24,25,26,27. While nanoparticles have long been used specifically as antimicrobial carriers, the use of nanoparticle-based carriers in polytherapy treatments with antibiotics in order to overcome antimicrobial resistance has been overlooked. Lyotropic liquid-crystalline nanoparticles such as cubosomes have been used as vehicles to deliver antibiotics into the cell28,29,30 and disrupt the membrane integrity of LPS-deficient bacteria (A. baumannii 19606R and A. baumannii 5075D) by solubilizing the OM31. In this study, we hypothesized that the combination of cubosomes with an antibiotic such as PMB which also destabilizes the OM might serve as a new strategy to combat antimicrobial-resistant bacteria.
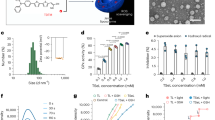
We selected four key antibiotics that are commonly used against Gram-negative bacteria, but with different modes of action (amikacin, aztreonam, doripenem and PMB) for investigation with A. baumannii, P. aeruginosa, and K. pneumoniae (Fig. 1a). The in vitro results showed that only membrane-targeting PMB in polytherapy with cubosomes enhanced bacterial killing against each of the examined pathogens. Subsequently, the antibacterial activity of PMB-loaded cubosomes was compared with the polytherapy treatment of PMB and cubosomes (Fig. 1b), showing that the polytherapy clinically relevant concentrations of PMB and cubosomes is superior in bacterial killing to PMB-loaded cubosomes. Confocal microscopy and neutron reflectometry (NR) were subsequently employed to investigate the detailed interactions of PMB/cubosomes with a model bacterial OM and thus determine the mechanism underpinning enhanced antibacterial activity. To the best of our knowledge, our study is the first to investigate the effect on bacterial killing via polytherapy with an antibiotic and a lyotropic liquid-crystalline lipid-based nanoparticle carrier against Gram-negative pathogens. Our findings offer novel insights that will assist in the design of new nanoparticles and therapies to act as potential adjuvants with existing and emerging antibiotics.
a Schematic representation of the OM bilayer exposed to different antibiotics which are commonly used in the treatment of Gram-negative bacteria. Amikacin irreversibly binds to the 30S subunit of bacterial ribosomes, interfering with mRNA binding and tRNA acceptor sites, thereby blocking protein synthesis and inhibiting bacterial growth; Aztreonam inhibits the synthesis of the bacterial cell wall by blocking peptidoglycan crosslinking. Doripenem inactivates PBPs by forming stable acyl-enzymes, resulting in a weakened cell wall; PMB interacts with lipopolysaccharide (LPS) of the OM of Gram-negative bacteria, causing the leakage of cytoplasmic content and finally cell death. b Schematic diagram of PMB-loaded cubosomes and the polytherapy treatment interacting with the Gram-negative bacterial OM. OM outer membrane, IM inner membrane, PBPs penicillin-binding proteins, mRNA messenger ribonucleic acid, tRNA transfer ribonucleic acid, PMB polymyxin B.
Results and discussion
Characterization of cubosomes
Dynamic light scattering (DLS) showed that cubosomes, Octadecyl Rhodamine B Chloride (R18)-loaded and 1–20 wt% PMB-loaded cubosomes had a size range of 150–170 nm and a zeta potential from −23.8 ± 1.0 mV to 1.6 ± 0.2 mV (Supplementary Fig. 1 and Supplementary Table 1). SAXS results show that a 1 wt% PMB loading in the cubosomes was found to be biphasic (QIID + QIIG cubic phases; Supplementary Figs. 2 and 3), while 2–20 wt% PMB-loadings resulted in a pure QIIG cubic phase, suggesting that increasing the amount of positively charged PMB within the cubosomes facilitates a transition from the QIID to the QIIG phase. It is noteworthy that in previous studies, the addition of additives such as charged lipids or nanoparticles also led to the stabilization of the QIIG phase in excess water29,32,33,34,35. In addition, the cubosome unit cell parameter and water channel radius expansion increased from 6.51 to 11.12 nm, and from 1.15 to 1.36 nm, respectively, which is driven by an increased electrostatic repulsion of the charged PMB within the cubic-phase nanoparticles (Supplementary Table 2). Cryogenic transmission electron microscopy also revealed the internally ordered structures of the PMB-loaded cubosomes (Supplementary Fig. 4).
The PMB entrapment efficiency (EE%) in 1–20 wt% PMB-loaded cubosomes was found to be higher than 94 % (Supplementary Table 3), indicating that most of the PMB was entrapped within the cubosomes. In vitro release profiles of the PMB-loaded cubosomes (Supplementary Fig. 5) showed that the rate and extent of PMB release increased with PMB concentration. Cubosomes loaded with 1, 2, and 4 wt% PMB released PMB slowly and steadily across the first 6 days, with a cumulative PMB release at this time of 37%, 49%, and 58%, respectively; no further PMB release occurred after this time. Increasing the PMB loading by 8, 16, and 20 wt%, results in a faster release across the first ~18 h, followed by a more gradual release prior to a plateauing. With these higher loading amounts, the cumulative release of PMB was 74%, 85%, and 97%, respectively. The initial faster release of PMB could be due to the rigid QIIG cubic phase containing less aqueous channels than the QIID cubic phase36, as well as electrostatic repulsion between the charged PMB molecules acting as a driving force for diffusion.
In vitro antimicrobial properties of the polytherapy treatment of antibiotic-cubosomes and PMB-loaded cubosomes
The minimum inhibitory concentration (MIC) was used to evaluate the antibacterial activity of amikacin, aztreonam, doripenem, and PMB, alone against representative isolates of A. baumannii, P. aeruginosa, and K. pneumoniae in cation-adjusted Mueller–Hinton broth (CaMHB) medium (Table 1). The cubosomes alone were ineffective against all bacterial strains (MICs > 32 µg/mL). The synergistic bacterial killing was measured via fractional inhibitory concentration (FIC) studies in CaMHB (Fig. 2a, c–i, black and red filled bars). The addition of cubosomes did not enhance the efficacy of amikacin, aztreonam, or doripenem (FIC index (FICI) > 1) in all seven strains. Importantly, however, cubosome polytherapy with the LPS targeting antibiotic, PMB, resulted in synergistic bactericidal activity against four of seven isolates (FICI ≤ 0.5) and partial synergy against the other three isolates (0.5 < FICI ≤ 1.0).
a Schematic representation of the FIC synergy checkerboard assay. b Schematic representation of the MIC assay. c–i FIC of antibiotic/cubosome polytherapy and MIC of (w/w %) PMB-loaded cubosomes against isolates of (c–f) A. baumannii, (g) P. aeruginosa, and (h, i) K. pneumoniae in CaMHB medium. Black and red filled bars represent antibiotics and cubosomes (Cub) in FIC, respectively; Black and red pattern bars represent the loaded PMB and the corresponding Cub in MIC, respectively. All data are expressed as the mean ± SD. All experiments were performed in triplicate (n = 3). Mean values and error bars were defined as the mean and SD, respectively. See Source Data for detailed FIC, FICI, and MIC values. MIC minimum inhibitory concentration, FIC fractional inhibitory concentration, Ami amikacin, Azt aztreonam, Dor doripenem, PMB polymyxin B. Source data are provided as a Source Data file.
We also explored the use of the traditional approach, namely the drug-loaded system, PMB-loaded cubosomes, against the same Gram-negative bacteria cohort (Fig. 2b, c–i, black and red pattern bars). Bacterial killing with all PMB-loaded cubosomes (1% to 20 wt% PMB) was substantially reduced against six of seven isolates when compared to the polytherapy approach as evidenced by the higher MICs (Fig. 2, black pattern bars). In contrast, the PMB-loaded cubosomes slightly lowered the concentration of PMB needed to inhibit the growth of A. baumannii FADDI-AB241, as evidenced by their lower MICs, but at the cost of using a high concentration of cubosomes (>32 µg/mL, Fig. 2f, red pattern bars). It is worth noting that 20 wt% PMB-loaded cubosomes with a significantly higher concentration of PMB in the system (~3.78 mg loading PMB, Supplementary Table 3) did not enhance the killing effect compared to the polytherapy approach. We further tested the bacterial killing with the polytherapy of PMB with 8 wt% PMB-loaded cubosomes (Supplementary Table 4). The analogous FIC values showed that PMB with 8 wt% PMB-loaded cubosomes did not significantly enhance their activities compared to the polytherapy approach (PMB with cubosomes) although there was more PMB in the bulk solution. These data suggest that the antimicrobials activity is not a PMB concentration dependant effect. The existence of cubosomes in polytherapy plays a crucial role in bacterial killing.
In order to mimic the in vivo conditions, MIC and FIC experiments were also performed in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10 v/v % fetal bovine serum (FBS) (Supplementary Data 1), which is a defined medium for mammalian cells. These results indicated that PMB-loaded cubosomes were more effective against the corresponding bacteria in DMEM/10% FBS than in CaMHB, yet still less potent compared to PMB/cubosome polytherapy in the same culture medium. In addition, the cell viability of a HEK-293T cell line following the addition of polytherapy indicated that there is no appreciable cytotoxicity on the studied mammalian cells up to 32 μg/mL (Supplementary Fig. 6). Notably, recent studies have shown for phytantriol cubosomes with a similar toxicity profile in vitro they were well tolerated in vivo37,38,39,40, which further highlights the potential for use of the polytherapy in vivo. In short, PMB-loaded cubosomes were less active than the polytherapy treatment in inhibiting the growth of Gram-negative pathogens. These data suggest that for lyotropic liquid-crystalline nanoparticles a “one-size-fits-all” approach, i.e., focusing only on conventional drug-loaded nanoparticles, is not sufficient for the identification of optimal regimens that maximize bacterial killing.
Interaction of cubosomes with bacteria visualized by confocal microscopy
Confocal microscopy was conducted in order to visualize the interactions of cubosome formulations with the bacteria. A. baumannii ATCC 19606 was selected as a suitable representative of the Gram-negative bacteria under investigation and treated with 32 μg/mL unloaded cubosomes, 1, 2, 4, 8, 16, and 20 wt% PMB-loaded cubosomes (corresponding to 0.32, 0.64, 1.28, 2.56, 5.12, and 6.4 μg/mL of PMB, respectively, with 32 μg/mL of cubosomes, and a polytherapy combination (0.5 μg/mL of PMB and 32 μg/mL of cubosomes).
When treated with unloaded cubosomes (Fig. 3a), a small number of cubosomes (in red) bound to the cell membrane (in green). This suggests that despite their negative charges (−23.8 ± 1.0 mV, Supplementary Table 1), cubosomes can still interact with the negatively charged bacterial OM (typically −130 to −150 mV)41. The previous cell mimetic studies of LPS-deficient model cell membranes have demonstrated that a diffuse cubosome layer forms only on the surface of Gram-negative bacteria without penetrating into the bacterial cells OM31. The lack of observed cubosomes penetrating the membrane likely explains their ineffectiveness at killing all the bacterial isolates studied (MICs > 32 µg/mL; Table 1) when used as a monotherapy alone.
Cubosomes were labeled with Octadecyl Rhodamine B Chloride (in red), such that the interactions between bacteria and cubosomes would be visible through the colocalization of their individual fluorescence emission in the micrographs. The white arrows indicate the slight interaction of cubosomes or PMB-loaded cubosomes with bacteria. All the images are representative of experiments performed in triplicate (n = 3). A. baumannii ATCC 19606 treated with (a) 32 μg/mL cubosomes alone; b 1 wt% PMB-loaded cubosomes; c 8 wt% PMB-loaded cubosomes; d 20 wt% PMB-loaded cubosomes and e polytherapy treatment with 32 μg/mL unloaded cubosomes and 0.5 μg/mL of PMB. Left: bacteria in green; middle: cubosomes in red; right: overlay image. Scale bar: 4 μm.
The binding of PMB-loaded cubosomes to the bacterial cell membrane was similar to that in the absence of PMB loading (Fig. 3b–d and Supplementary Fig. 7), despite the presence of positively charged PMB in the cubosomes. Surprisingly, a majority of cells following polytherapy treatment had yellow fluorescence after incubation, indicating increased binding of cubosomes to the outside of the bacterial membrane compared to that of the unloaded- and PMB-loaded cubosomes (Fig. 3e). Significantly, PMB is well-known for its ability to interact electrostatically with, and subsequently disrupt, the OM of Gram-negative bacteria42, while cubosomes are known to cause structural rearrangement of the inner leaflet of the OM31. Thus, the enhanced antibacterial activity observed with the polytherapy approach (Fig. 2) is likely due to synergistic interactions of both PMB and the cubosomes with the bacterial OM. To understand the possible mechanisms of these interactions, we employed the use of NR on a model bacterial cell membrane which is reported next.
Interactions of a polytherapy treatment with a model Gram-negative bacterial OM via NR
NR is a state-of-the-art tool for non-destructively probing a model bacterial membrane and discriminating between different membrane components with a spatial resolution approaching a few Ångström43. Importantly, we have previously demonstrated using NR that cubosomes alone bind only weakly to both a model Gram-negative bacterial OM composed of Ra-LPS, which contains the lipid A and core oligosaccharide region of LPS, and a model inner membrane, but were unable to penetrate into the center of either membrane31. By excluding the Ra-LPS and inner membrane, we specifically focused on the hydrophobic domain of LPS, i.e., lipid A. NR was undertaken to investigate the interaction between the components of a polytherapy treatment and a model OM of Gram-negative bacteria consisting of lipid A in the outer leaflet and 1,2-dipalmitoyl-d62-sn-glycero-3-phosphocholine (d-DPPC) in the inner leaflet (Fig. 4a). Due to the membrane-targeting action of both PMB and cubosomes, each component has the potential to initially interact with the bacterial membrane surface. Thus, to determine the bactericidal mechanism of the polytherapy treatment, two experiments were conducted whereby the interaction sequence between cubosomes and PMB with the bacterial OM were reversed.
a Schematic representation of the model OM bilayer. The lipid A (outer leaflet): d-DPPC (inner leaflet) membrane bilayer are absorbed on the SiO2 surface. During experiments, all space inside the cell is always filled with the aqueous solution, i.e., D2O, CMSi (contrast-matched silicon), and H2O buffer. b The NR profiles (symbols) and fits (lines) of the bilayer obtained from D2O (black squares and line), CMSi (red circles and line), and H2O (blue up triangle and line), respectively. The insert is the corresponding scattering length density (SLD) profiles of the bilayer whereby the cartoon depicts the bilayer structure and the distance relative to the SiO2 surface. c Experimental (symbols) and fitted (curves) NR profiles of the bilayer (black square and line), treated with 32 µg/mL cubosomes (red circle and line) followed by 4 µg/mL PMB (blue up triangle and line) in D2O; black arrow indicates the fringe changing from black to red curve after incubation with cubosomes. d The corresponding SLD profiles of (c). The inset depicts the membrane bilayer after successive treatment with cubosomes and PMB. e Experimental (symbols) and fitted (curves) NR profiles of the bilayer (black square and line) treated with 4 µg/mL PMB (red circle and line) followed by 32 µg/mL cubosomes (blue up triangle and line) in D2O; black arrow indicates the fringe from black to red curve remained the same after incubation with PMB. f The corresponding SLD profiles of (e). The insert depicts the bilayer after successive treatment with PMB and cubosomes.
OM of Gram-negative bacteria—cubosome treatment followed by PMB treatment
The bilayer was constructed using the Langmuir−Blodgett and Langmuir−Schaefer techniques (Supplementary Fig. 8a, b, see “Methods” for further clarification)44,45 before being subjected to NR measurement (Supplementary Fig. 8c). The NR profiles of the OM bilayer were then obtained as shown in Fig. 4b. Each isotopic contrast that the OM bilayer was measured under was modeled simultaneously to accurately depict the structure of the asymmetric OM (see Supplementary Fig. 9 for further clarification). The lipid volume fractions across the SiO2 surface for the bilayer were calculated to be as high as 95.3 ± 3.0% (Supplementary Table 5), indicative of the successful fabrication of the OM of Gram-negative bacteria.
Following its construction, the model OM was treated with 32 µg/ml cubosomes. A flattened fringe was clearly observed in the reflectivity curve (Fig. 4c, arrow), indicating OM perturbation. In addition, a decreased scattering length density (SLD) in the inner tail region (Fig. 4d, from black to red curves) was observed. The fitted parameters (Supplementary Table 6) confirmed the presence of a diffuse phytantriol layer (thickness of 50.7 ± 0.1 Å) adsorbed to the surface of the lipid A headgroups of the outer leaflet. Subsequently, a small volume fraction of phytantriol (~6%, Supplementary Table 6) was detected in the inner and outer leaflets of the OM. The NR results suggest that cubosomes can interact with lipid A and then partly penetrate into the phospholipid layer. However, the bilayer remained highly intact (~81%) as only a small percentage of phytantriol was found in the bilayer.
With the membrane having initially been exposed to the cubosomes, further treatment with 4 µg/mL PMB was undertaken. Following the addition of PMB, there was only a negligible shift in reflectivity profiles (Fig. 4c, from red to blue curve). At the same time, the SLD in both leaflets remained the same (Fig. 4d), implying that the addition of PMB to the cubosome-treated membrane did not induce significant changes to the bilayer structure. The best fit parameters (see Supplementary Table 7) revealed that PMB was prevented from entering the bilayer as indicated by the absence of PMB in the sublayers. Consequently, the volume fractions of the lipids in the bilayer remained very high (>80.6%). Notably, negligible absorption of PMB or phytantriol cubosomes was observed on the silica surface according to analysis via Quartz crystal microbalance with dissipation (QCM-D) responses (Supplementary Fig. 10)46. This suggests that the NR experimental results were not adversely influenced from the silica support. Taken together, NR results suggest that a diffuse cubosome layer accumulated on the membrane but phytantriol did not exchange with the lipids within the bilayers to any significant extent to cause further damage. Further incubation with PMB only led to the PMB adsorbing onto the outer cubosome layer without penetrating into the membrane structure. Thus, PMB was prevented from reaching the OM due to blockage by the cubosomes which overlaid the OM surface. The lack of membrane damage associated with PMB-loaded cubosomes agrees with the in vitro results (Fig. 2).
OM of Gram-negative bacteria—PMB treatment followed by cubosome treatment
Similarly, as to the prior study, the NR data confirmed the presence of the membrane bilayer with a high-volume fraction of lipid of 98.1 ± 3.5% across the SiO2 surface (Supplementary Fig. 11 and Supplementary Table 8). The bilayer was initially treated with 4 μg/mL PMB, followed by a 32 μg/mL cubosome treatment (Supplementary Fig. 11c–f).
In good agreement with our previous study45, the fringe in the NR curve and the SLD in the tail region remained unchanged (Fig. 4e arrow and Fig. 4f, from black to red curve) following PMB treatment, and a negligible amount of PMB (volume fraction ~2.7–4.8%) was detected in the OM sublayers (Supplementary Table 9). It is thus unlikely that incubation with PMB significantly compromised the membrane structure. We, therefore, propose that PMB primarily binds to the OM surface without extensive penetration into the lipid bilayer structure at a bulk concentration of 4 μg/mL. Our previous study using a higher concentration of PMB (16 μg/mL) revealed that approximately a volume fraction of 10% of PMB binds to the fatty acyl region of lipid A. This indicates hydrophobic interactions are occurring between PMB and lipid A in addition to the primary electrostatic interactions44. The low PMB penetration observed in both experiments is possibly due to the stable gel-like state of the phytantriol bilayer structure at 25 °C. Whereas, at higher temperatures, the bilayer can change phases from a gel to a disordered liquid-crystalline state and enhance PMB penetration47,48. Following subsequent treatment with 32 μg/mL cubosomes, significant changes in the fringe of the NR curve (Fig. 4e, from red to blue curves) and in the tail regions of SLD (Fig. 4f) were observed, indicating solubilization of the bilayer following polytherapy (Fig. 4f, blue curve). Analysis of the NR profiles indicated that large volume fractions of phytantriol (up to 20.3 ± 0.5%) and water (up to 13.4 ± 6.5%) were found in the sublayer (Supplementary Table 10). As a result, PMB with volume fractions ranging from 6.7 ± 0.9% to 9.7 ± 0.5% was detected across the bilayer structure, which is possibly due to electrostatic and hydrophobic interactions between the amphipathic PMB and lipids in the sublayers42,44. Consequently, a drastic decrease in the volume fraction (from 98.1 ± 3.5% to ~65.6 ± 0.4%) of the bilayer was observed (Supplementary Table 10) which accords with the flattened fringe. Thus, the PMB greatly enhanced the interaction of cubosomes with the model bacterial OM.
Proposed bactericidal mechanism of the polytherapy treatment of PMB and cubosomes against Gram-negative bacteria
We have previously reported that cubosomes alone are unable to disrupt the OM of Gram-negative bacteria31. Based on the confocal microscopy (Fig. 3) and NR (Fig. 4) results, Fig. 5 illustrates the proposed antimicrobial mechanisms by which the polytherapy treatment disrupted the OM of Gram-negative bacteria. In Fig. 5a, PMB-loaded cubosomes attach to the bacterial membrane surface without penetrating it. The adsorbed layer of cubosomes subsequently prevented the released PMB from binding to the membrane, thus making it more impervious to PMB attack. Consequently, the membrane remained relatively undamaged. Figure 5b describes two distinct modes of action of the polytherapy treatment on the Gram-negative bacterial model OM. First, membrane stability is initially compromised by PMB binding to the membrane surface due to electrostatic interactions between the polycations on PMB and the negatively charged phosphate groups on lipid A49,50,51. Second, the entry of cubosomes into the bilayer is facilitated due to water influx through the partially permeabilized membrane. Cubosomes are then expected to interact with the bilayer lipids via a lipid-exchange mechanism31, whereby the membrane bilayer is further solvated. This finding indicates that enhanced bacterial killing of Gram-negative bacteria by polytherapy is not contingent upon combining two antimicrobials with separate antimicrobial activity.
Treatment with (a) PMB-loaded cubosomes and (b) PMB combined with cubosomes. Polytherapy with PMB and cubosomes result in interactions with the bacterial OM in two consecutive ways: PMB initially interacts with the outer leaflet of OM via electrostatic interactions, leading to destabilized areas. Cubosomes then contact with the bilayer, causing further membrane perturbations via a lipid-exchange process.
In summary, we have demonstrated that cubosomes in a polytherapy or combination approach with the membrane-targeting antibiotic PMB significantly enhance bacterial killing of problematic Gram-negative bacteria, including polymyxin-resistant strains. The PMB-loaded cubosomes, however, did not express a better synergistic effect with PMB compared to cubosomes. Strong interactions between the components during a polytherapy treatment with bacterial OM’s were observed by confocal microscopy, yet rarely observed for PMB-loaded cubosomes. Mechanistic studies employing a model cell membrane and NR suggested PMB initially destabilized the Gram-negative OM followed by cubosome-induced membrane solubilization via a lipid-exchange process. These findings pave the way for the future design of nanoparticle-based combination therapies that can potentially overcome Gram-negative bacterial resistance to antimicrobials.
Methods
Antibiotics, reagents, and cell line
Polymyxin B (PMB; lot number 20120204), amikacin (lot number 058K0803), aztreonam (batch number MKCH8931), and doripenem (lot number 0137Y01) were purchased from Sigma-Aldrich and their solutions were prepared in sterilized Milli-Q water (Millipore, Australia). Phytantriol (98%, 3,7,11,15-tetramethylhexadecane-1,2,3-triol), poly(ethylene oxide)-poly(propylene oxide)-poly(ethylene oxide) triblock copolymer (Pluronic F127), lipid A (diphosphoryl from Escherichia coli F583), Octadecyl Rhodamine B Chloride (R18), Dulbecco’s modified Eagle’s medium (DMEM, 4.5 g/L glucose), fetal bovine serum (FBS) and [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (tetrazole)] (MTT) were purchased from Sigma-Aldrich; 1,2-dipalmitoyl-d62-sn-glycero-3-phosphocholine (d-DPPC) was purchased in powder form from Avanti Polar Lipids Inc. CellBriteTM Fix 488 membrane stain was purchased from Biotium. Milli-Q water 18.2 MΩ ∙ cm and D2O (99.99 atom%) were used in all experiments for preparation of buffers and solutions. All chemicals were used as received without further purification. The Human embryonic kidney 293T cells (ATCC HEK-293T) was the gift from Dr Thomas Naderer, Monash University.
Preparation of cubosome formulations
Eight types of phytantriol based nanoparticles were prepared: phytantriol cubosomes; 0.05 wt% R18-99.95 wt% phytantriol (R18-labeled cubosomes); 1 wt% PMB-99 wt% phytantriol (1 wt% PMB-loaded cubosomes); 2 wt% PMB-98 wt% phytantriol (2% PMB-loaded cubosomes); 4 wt% PMB-96 wt% phytantriol (4% PMB-loaded cubosomes); 8 wt% PMB-92 wt% phytantriol (8% PMB-loaded cubosomes); 16 wt% PMB-84 wt% phytantriol (16% PMB-loaded cubosomes); 20 wt% PMB-80 wt% phytantriol (20% PMB-loaded cubosomes); Phytantriol cubosomes were prepared by weighing phytantriol in a glass vial followed by adding a solution of Pluronic F127 in chloroform at a 10 wt% to the phytantriol as previously described31,52. The chloroform in the mixture was dried using a nitrogen gas dryer, followed by vacuum drying at room temperature overnight to obtain a viscous and transparent gel. Milli-Q water/or PBS buffer were then added to the glass vial prior to dispersion by ultrasonication (Qsonica sonicators, Adelab Scientific) for 5 mins in pulse mode (5 s pulses interrupted by 5 s breaks) at 50% of maximum power (125 Watt, 20 kHz) in ice bath to give a milky dispersion. Finally, the glass vial was sealed and kept at room temperature for further characterization. R18-labeled cubosomes were obtained by adding R18 to the phytantriol prior to Pluronic F127 addition. For PMB-loaded nanoparticles, Pluronic F127 at a 10 wt% of total phytantriol and PMB was added to the phytantriol. After overnight drying, the vial was placed in a 70 °C water bath until the gel was melted, 4 mg of PMB dissolved in 50 µL of sterilized Milli-Q water/or PBS buffer were then added to the vial and thoroughly mixed with the melted gel by vortex and sonication to accomplish a state of homogeneity at room temperature. The final PMB concentration was defined as the actual loaded amount of PMB after removal of the free PMB in the final dispersion.
In vitro PMB entrapment and release studies
To separate the free PMB from the cubosome dispersions, ultrafiltration centrifugation was performed prior to the entrapment efficiency determination. 0.5 mL of PMB-loaded cubosomes were filtered through an Amicon®Ultra – 0.5 mL centrifugal filter (Merck Millipore Ltd.) with a regenerated cellulose 10,000 Nominal Molecular Weight Limit (NMWL) at 14,000×g for 30 min. The free PMB in the filtrate was collected for further measurement. Subsequently, the filters were placed upside down in a fresh Eppendorf tube and centrifuged at 5000×g for 10 min to recover the PMB-loaded cubosomes.
In vitro release profiles of PMB from cubosomes were evaluated using a dynamic dialysis method53. Briefly, the recovered PMB-loaded cubosomic dispersions were loaded into a dialysis bag (14,000 MWCO, Millipore, Boston, USA) and dialyzed against 20 mL of phosphate buffer saline (PBS, pH 7.4; the concentration of Na2HPO4, KH2PO4, NaCl, and KCl were 10, 1.8, 137, and 2.7 mmol/L, respectively) thermostatically maintained (37 ± 0.5 °C) and magnetically stirred (200 rpm) throughout the experiment. At 0.5, 1, 2, 4, 18 h, 1d, and then daily up until 12d, 1 mL of solution was removed and replaced by 1 mL of fresh PBS buffer to confirm sink conditions.
Utilizing ultraviolet/visible (UV/Vis) measurement (SparkControl V2.3, Spark 10 M), the amounts of free PMB and the released PMB from the dialysis bag were determined at 210 nm and then equated with standard curve fitting. All the studies and measurements were conducted in triplicate. All the data were processed by Microsoft 365 version 2105 and Originlab 2018 b9.5.1.195.
The percentage of the PMB entrapped in the cubosomes was calculated as: PMB entrapment efficiency (EE%) = (Mtotal − Mfree)/Mtotal * 100%, where Mtotal is the total PMB added in the cubosomes, and Mfree is the free PMB in the filtrate after ultrafiltration.
Dynamic light scattering
The particle size distribution, polydispersity index (PDI) along with the Zeta potential (ζ) of cubosomes were measured on Malvern Zetasizer nanoZS (Zetasizer Software V6.28, Monash Biomedicine Discovery Institute). Samples were diluted with Milli-Q water to a concentration of 0.1 mg/mL and injected into DTS 1070 folded capillary cells. The samples in the cells were stabilized at 37 °C and the DLS was recorded in triplicate at 173° (backscattering angle) for 60 s. ζ -potentials were recorded 30 runs in triplicate.
Small-angle X-ray scattering (SAXS)
SAXS was performed at 25 °C using a compact Bruker N8 Horizon instrument (Using software Diffrac. Suite V7.3.1, Monash X-ray Platform, Monash University, Australia), which allows irradiation of samples within a sealed X-ray tube HV generator (Kα radiation from Cu-anode, wavelength λ = 1.5406 Å) at 50 keV and 1000 μA. The investigated Q-range was from 0.007 to 0.387 Å−1 (scattering vector Q = [4π sin(θ/2)]/λ, where θ is the scattering angle). Cubosomes at a concentration of 100 mg/mL were then loaded into a 1 mm (inner diameter) quartz capillary with Milli-Q water as background and Glassy carbon as the reference for determining sample transmission. The sample chamber was kept under vacuum during the measurement. The 2D scattering pattern was then recorded by VÅNTEC-500 Detector for 10 h. The image was then integrated into the 1D scattering function I(Q) and the background of Milli-Q water or PBS buffer in the capillary was subtracted by DIFFRAC.SAXS software (Monash X-ray Platform, Monash University, Australia). A silver behenate standard with a d spacing value of 58.38 Å was used for calibration. The cubic lattice parameter a was calculated as: a = \({d}_{{hkl}}\sqrt{\left({h}^{2}+{k}^{2}+{l}^{2}\right)}\), where the lattice spacing dhkl = 2π/Q, h, k, and l are miller indices. The water channel radius of the cubic structure was estimated by rw = [(−σ/2πχ)1/2a] − l, where σ and χ are topological constants (σ = 1.919 and χ = −2 for Pn3m structure; σ = 3.091 and χ = −8 for Ia3d structure), l is the length of the lipid chain (ca. 1.4 nm for phytantriol)54,55. The estimated rw equations for cubosomes are given as: rPn3mw = 0.391a − 1.4 and rIa3dw = 0.248a − 1.4.
Cryogenic transmission electron microscopy (Cryo-TEM)
A laboratory-built humidity-controlled vitrification system was used to prepare the samples for Cryo-TEM. Humidity was kept close to 80% for all experiments, and ambient temperature was 22 °C. In all, 300-mesh copper grids coated with perforated carbon film (Lacey carbon film: ProSciTech #GSCU300FL-50, QLD, Australia) were glow discharged to render them hydrophilic. In total, 3 μl aliquots of the sample were pipetted onto each grid prior to plunging. After 5 s adsorption time the grid was blotted manually using Whatman 541 filter paper, for ~2 s. The grid was then plunged into liquid ethane cooled by liquid nitrogen. Frozen grids were stored in liquid nitrogen until required. The samples were examined using a Gatan 626 cryoholder (Gatan, Pleasanton, CA, USA) and Tecnai 12 Transmission Electron Microscope (FEI, Eindhoven, The Netherlands) at an operating voltage of 120KV. At all times, low-dose procedures were followed, using an electron dose of 8–10 electrons/Å2 for all imaging. Images were recorded using a FEI Eagle 4k x 4k CCD camera at a range of magnifications using AnalySIS v3.2 camera control software (Olympus).
Minimum inhibitory concentration (MIC) testing
The MIC is used to determine the lowest concentration of a drug that prevents the visible growth of a bacterium or bacteria. MIC testing was performed according to Clinical and Laboratory Standards Institute (CLSI) guidelines using amikacin, aztreonam, doripenem, and PMB56,57. A colony-forming unit (CFU) is used to estimate the number of viable bacteria in a sample. Briefly, twofold dilutions of the antimicrobial agent were prepared in Cation-Adjusted Mueller–Hinton Broth (CaMHB, 20–25 mg/L of Ca2+, 10–12.5 mg/L of Mg2+ in Mueller–Hinton Broth, a nutrient-rich bacterial growth medium), or in Dulbecco’s modified Eagle’s medium (DMEM) containing 10 v/v % fetal bovine serum (FBS). A 96-well microtiter plate was firstly filled with 100 µL of increasing antimicrobial concentrations. The bacterial suspension containing either representative strains of Acinetobacter baumannii, Pseudomonas aeruginosa or Klebsiella pneumoniae was then standardized to 0.5 using a McFarland meter giving ~108 CFU/mL of the bacteria. The suspension was then diluted 1:100 with CaMHB to achieve ~106 CFU/mL of the viable bacteria. Within 15 min, 100 µL of the diluted suspensions were vortexed and pipetted into the above wells, resulting the final inoculum of ~5 × 105 CFU/mL. The MIC was determined after incubation at 37 °C for 16–20 h. The experiment was performed in duplicate for each antibiotic and bacterium. All the data were processed by Microsoft 365 version 2105 and GraphPad Prism version 9.0.1 (151).
Susceptibility and resistance were determined according to the European Committee of Antimicrobial Susceptibility Testing (EUCAST) guidelines58. As only breakpoints for colistin (not polymyxin B) against A. baumannii, P. aeruginosa and K. pneumonia have been established by EUCAST and given colistin and polymyxin B have essentially identical in vitro potencies as measured by MICs59, the EUCAST breakpoints for colistin were applied to polymyxin B. Susceptibility (S) and resistance (R) were defined as MICs ≤2 and >2 µg/mL for polymyxin B, respectively, against all bacterial species tested. For amikacin, S and R were defined as ≤16 and >16 µg/mL against P. aeruginosa, and ≤8 and >8 µg/mL for A. baumannii and K. pneumonia. For aztreonam, S and R were defined as ≤0.001 and >16 µg/mL for P. aeruginosa and ≤1 and >4 µg/mL L for K. pneumonia; breakpoints for aztreonam against A. baumannii have not been established. For doripenem, S and R were defined as ≤0.001 and >2 µg/mL for A. baumannii and P. aeruginosa, and ≤1 and >2 µg/mL for K. pneumonia.
Fractional inhibitory concentration (FIC) testing
FIC is used to determine the impact on the potency of an antibiotic combination in comparison to the individual activities of each antibiotic. FIC testing was undertaken using the microdilution checkerboard technique according to CLSI guidelines60. Briefly, twofold dilutions of solutions (four antibiotics, i.e., amikacin, aztreonam, doripenem, and PMB in combination with cubosomes, respectively) were performed in CaMHB. A 96-well microtiter plate was then filled with 50 µL of the appropriate concentrations of antibiotic solution, and this step repeated with cubosome solution (total final volume, 100 µL). A bacterial suspension containing either A. baumannii, P. aeruginosa or K. pneumoniae was then diluted with CaMHB to give ~106 CFU/mL of final suspension, with 100 µL then dispensed into the wells (final inoculum, ~5 × 105 CFU/mL). The FIC was determined after incubation at 37 °C for 16–20 h. The experiment was performed in triplicate for each antibiotic and bacterium. All the data were processed by Microsoft 365 version 2105 and GraphPad Prism version 9.0.1 (151).
The FIC index (FICI) is the lowest concentration of antimicrobial agents showing complete inhibition of growth as detected by the unaided eyes. It is calculated by the equation: FICI = (FICA1/MICA1)/(FICA2/MICA2), where A1 = Antibiotic 1, A2 = Antibiotic 2. Synergy was defined as FICI ≤ 0.5, additivity (partial synergy) as 0.5 < FICI ≤ 1.0, indifference as 1.0 < FICI < 4.0, and antagonism as FICI ≥ 4.061. If the endpoint MIC value was unable to be determined due to growth at the highest concentration used, then the next MIC value was used for the calculation (e.g., for an MIC > 32 µg/mL, the MIC used for the calculation was 64 µg/mL)20.
Cell viability test
The cell viability was examined using MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (tetrazole)] assay. Human embryonic kidney cells (HEK293) were seeded into tissue culture-treated 96-well plates at 5 × 103 cells per well and incubated at 37 °C in the presence of 5% CO2 overnight. The cells were treated with culture medium containing different concentrations of cubosomes, PMB and PMB/cubosomes overnight. Afterward, the medium was replaced with fresh medium containing 0.5 mg/mL MTT and incubated at 37 °C for 4 h. When finished, the medium was replaced by DMSO and incubated at 37 °C for 10 min. Tecan M200 plate reader (DKSH, Australia) was used to measure the absorbance of the solution at 570 nm.
Confocal microscopy
A. baumannii ATCC 19606 was used for the microscopy. The bacterium was streaked on CaMHB agar plates and cultured at 37 °C overnight. One colony from the plate was transferred to 10 mL of CaMHB and grown overnight under shaking conditions (200 rpm) at 37 °C. The overnight suspension was then diluted in CaMHB to reach a final concentration of ~108 CFU/mL as measured by OD600 0.5. Bacterial suspensions (1 mL) were then incubated with the CellBriteTM Fix 488 membrane stain at 37 °C for 15 min and washed thrice with phosphate-buffered saline (PBS). The stained bacterial suspension was centrifuged at 3000×g and the pellets obtained were resuspended in 1 mL of CaMHB. Samples (PMB, R18-labeled cubosomes, R18-labeled PMB-loaded cubosomes) at their MIC values (0.5 µg/mL PMB; for polytherapy, the R18-labeled cubosomes had a fixed concentration of 32 µg/mL; For R18-labeled PMB-loaded cubosomes, 1, 2, 4, 8, 16, and 20 wt% PMB-loaded cubosomes have 0.32, 0.64, 1.28, 2.56, 5.12, and 6.4 μg/mL of PMB, respectively, with 32 μg/mL of cubosomes) were then added to the suspensions respectively and incubated for 2 h at 37 °C under shaking conditions (200 rpm). Bacteria were mounted on a glass microscope slide, anti-fade mounting media, and glass coverslip. Images were acquired using a LEICA-SP5 confocal microscope (Leica Microsystems. Mannheim, Germany) equipped with a ×100 objective (NA: 1.40, oil) lens at Monash MicroImaging, Monash University, Australia. All the data were processed by ImageJ 1.53a.
Asymmetric bilayer deposition for NR
Lipid A and d-DPPC were used in this study to mimic the outer and inner leaflets of the Gram-negative outer membrane (OM), respectively. The asymmetric OM bilayer was formed using the Langmuir−Blodgett (LB) and Langmuir−Schaefer (LS) techniques (Supplementary Fig. S1)31,44,45, whereby a precise control over the components and conformation of each leaflet of lipid bilayer was achieved.
NR measurement
NR measurements were conducted on the Platypus time-of-flight neutron reflectometer at the 20 MW OPAL research reactor at the Australian Nuclear Science and Technology Organization (ANSTO), Sydney, Australia62, using a cold neutron spectrum 2.8 Å ≤ λ ≤ 18 Å. A chopper pairing of choppers 1 and 4 set to a 24 Hz rotation speed was used and provides a wavelength resolution (Δλ/λ) ~8%. Using a vertical scattering geometry, the neutron beams reflected from the sample interface were collected at two glancing angles of incidence (0.85° for 300 s and 3.5° for 3600 s) to cover the momentum transfer (Q) range of interest (0.01 Å−1 ≤ Q ≤ 0.3 Å−1), where Q is defined as: Q = 4π sin(θ)/λ, where θ is the angle of incidence and λ the wavelength. An illuminated footprint of 33 mm wide and 50 mm long was used.
The asymmetric membrane bilayer was characterized in three isotopic contrasts with contrast-matched scattering length density (SLD, ρ), i.e., D2O (99.9%, ρ = 6.35 × 10−6 Å−2), CMSi (contrast-matched silicon, 38% D2O: 62% H2O v/v; ρ = 2.07 × 10−6 Å−2) and H2O (ρ = −0.56 × 10−6 Å−2). For the exchange between these three contrasts, a total of 7 mL of pH/D 7.4 buffer solution (5 mM CaCl2, 150 mM NaCl, and 10 mM HEPES) was pumped through the sample wafer at the rate of 1.0 mL/min using a HPLC pump (Knauer GmbH, Berlin, Germany). Initially, the bilayer was characterized in buffered isotopic contrasts to confirm the successful formation of the model membrane before incubation for 4 h with the first treatment (32 µg/ml cubosomes or 4 µg/mL PMB), which was subsequently washed out by the injection of D2O buffer to remove any unspecific bindings. The model membrane after the first treatment was then fully characterized under the three isotopic contrasts. Finally, the second treatment (32 µg/ml cubosomes or 4 µg/mL PMB) was added in the same manner to the pre-treated membrane to obtain the final profiles of the model OM of Gram-negative bacteria after polytherapy.
Data reduction was conducted using the SLIM data reduction routine which accounts for detector efficiency, converts the time-of-flight data to wavelength, which is then used to calculate Q, re-bins the data to instrument resolution, stitches the datasets from the two angles of incidence at the overlap region to provide a complete reflectivity profile, and scales the data so that the critical edge is equal to a reflectivity of one.
NR data analysis
The analysis of NR profiles was carried out using the MOTOFIT analysis program in Igor pro 6.37 software (WaveMetrics Inc., Portland, OR)63. In this study, the bilayer was divided into several sublayers each defined by its thickness, SLD (Supplementary Table S1), and roughness. The reflectometry profiles of the bilayer under different isotopic contrasts (D2O, CMSi, and H2O) were then fitted simultaneously to determine the volume fractions of each component in the sublayers. When a layer is composed of two components, namely a chemical species s and water w, the resultant SLD can be given by: ρlayer = φρs + (1 − φ) ρw, where ρs and ρw are the SLD of two components (the SLDs used in this study are presented in Supplementary Table 11), respectively, and φ is the volume fraction of chemical species s in the layer. The relative volume fractions of the d-DPPC, lipid A, and PMB in the headgroup of the bilayer structure were unable to be determined because of the minimal isotopic contrast between these three headgroups. Likewise, the relative volume fractions of phytantriol and Lipid A in the tail region were unable to be determined64. The volume fractions are shown as a percentage and were determined by the combined volume fractions of the lipid tails region of the bilayer, as the hydration of lipid headgroup regions is more likely to be significantly higher than that of the tail regions owing to the hydrophilic nature of this moiety of the bilayer31.
Quartz crystal microbalance with dissipation monitoring (QCM-D)
QCM-D is widely used for the evaluation of the binding and interactions between the biological molecules and surfaces within liquid65. It was performed by employing Q-SENSE E4 system equipped with an axial flow chamber, IPC High Precision Peristaltic Multi-Channel Dispenser (ISMATEC, flow rate 0.2 mL/min), and 50 nm SiO2-coated quartz crystal sensor66 at 25 °C. Briefly, the sensors were cleaned by sodium dodecyl sulfate (SDS, 10 mM) for 8 h and then rinsed extensively with Milli-Q water, followed by nitrogen drying and oxidation in UV–ozone chamber for 15 min to remove residual organic impurities. Upon adsorption of materials to the surface of a sensor crystal, the shifts in resonance frequency (Δf, reflecting the mass of adsorbents, including the coupled water, on the sensor surface) and the changes in energy dissipation (Δd, correlating with the viscoelastic properties of molecular layers on the surface of the sensor) can be detected from QCM-D using the software QSoft401 V2.5.2.418. The resonance frequency and the dissipation were measured simultaneously at the fundamental natural frequencies 15 and 25 MHz, corresponding to the 3rd and 5th harmonics (overtones) of the 5 MHz crystal. The frequency change (Δf) of oscillating quartz could be linearly related to its mass change (Δm) as expressed by the Sauerbrey equation: Δm = −C * 1/n * Δf, where n is the overtone number and C is a constant, which is approximately equal to 17.7 ng/(cm2 · Hz) for a 5 MHz AT-cut quartz crystal at room temperature. This means that the addition of 17.7 ng/cm2 of mass on a 5 MHz quartz crystal causes a frequency change of 1 Hz. The frequency of 5 MHz quartz can be easily measured with a precision of 0.01 Hz in a vacuum. Therefore, measurement of nanogram-scale masses can be achieved. For example, the corresponding frequency shift on the addition of a monolayer of water with an areal density of ~25 ng/cm2 to the surface of an AT-cut quartz crystal is ~1.4 Hz and well within the limits of detection.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The authors declare that the data supporting the findings of this study are available within the article and its Supplementary files. Source data are provided with this paper.
References
Matsunaga, N. & Hayakawa, K. Estimating the impact of antimicrobial resistance. The Lancet Global Health 6, e934–e935 (2018).
Asokan, G. V., Ramadhan, T., Ahmed, E. & Sanad, H. WHO global priority pathogens list: a bibliometric analysis of Medline-PubMed for knowledge mobilization to infection prevention and control practices in Bahrain. Oman Med. J 34, 184 (2019).
Rojas, E. R. et al. The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 559, 617–621 (2018).
Santajit, S. & Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed. Res. Int. 2016, 2475067 (2016).
Zavascki, A. P., Goldani, L. Z., Li, J. & Nation, R. L. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother. 60, 1206–1215 (2007).
Velkov, T., Thompson, P. E., Azad, M. A., Roberts, K. D. & Bergen, P. J. History, chemistry and antibacterial spectrum. Polymyxin Antibiotics: From Laboratory Bench to Bedside 1145, 15–36 (2019).
Li, Z. & Velkov, T. Polymyxins: mode of action. Polymyxin Antibiotics: From Laboratory Bench to Bedside 1145, 37–54 (2019).
Giamarellou, H. Epidemiology of infections caused by polymyxin-resistant pathogens. Int. J. Antimicrob. Agents 48, 614–621 (2016).
Lesho, E. et al. Emergence of colistin-resistance in extremely drug-resistant Acinetobacter baumannii containing a novel pmrCAB operon during colistin therapy of wound infections. J. Infect. Dis 208, 1142–1151 (2013).
Lenhard, J. R., Nation, R. L. & Tsuji, B. T. Synergistic combinations of polymyxins. Int. J. Antimicrob. Agents 48, 607–613 (2016).
Isler, B., Doi, Y., Bonomo, R. A. & Paterson, D. L. New treatment options against carbapenem-resistant Acinetobacter baumannii infections. Antimicrob. Agents Chemother. 63, e01110–e01118 (2019).
Hussein, M. et al. Metabolomics study of the synergistic killing of polymyxin B in combination with amikacin against polymyxin-susceptible and-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 64, e01587-19 (2019).
Rao, G. G. et al. Polymyxin B in combination with doripenem against heteroresistant Acinetobacter baumannii: pharmacodynamics of new dosing strategies. J. Antimicrob. Chemother. 71, 3148–3156 (2016).
Lenhard, J. R. et al. Comparative pharmacodynamics of four different carbapenems in combination with polymyxin B against carbapenem-resistant Acinetobacter baumannii. Int. J. Antimicrob. Agents 48, 719–724 (2016).
Han, M. L. et al. Metabolic analyses revealed time-dependent synergistic killing by colistin and aztreonam combination against multidrug-resistant Acinetobacter baumannii. Front. Microbiol. 9, 2776 (2018).
Thomas, V. M., Brown, R. M., Ashcraft, D. S. & Pankey, G. A. Synergistic effect between nisin and polymyxin B against pandrug-resistant and extensively drug-resistant Acinetobacter baumannii. Int. J. Antimicrob. Agents 53, 663–668 (2019).
Lin, Y.-W. et al. Novel polymyxin combination with the antiretroviral zidovudine exerts synergistic killing against NDM-producing multidrug-resistant Klebsiella pneumoniae. Antimicrob. Agents Chemother. 63, e02176–18 (2019).
Tran, T. B. et al. Novel polymyxin combination with antineoplastic mitotane improved the bacterial killing against polymyxin-resistant multidrug-resistant Gram-negative pathogens. Front. Microbiol. 9, 721 (2018).
Krishnamurthy, M. et al. Enhancing the antibacterial activity of polymyxins using a nonantibiotic drug. Infect. Drug Resist. 12, 1393–1405 (2019).
Hussein, M. H. et al. From breast cancer to antimicrobial: combating extremely resistant Gram-negative “superbugs” using novel combinations of polymyxin B with selective estrogen receptor modulators. Microb. Drug Resist. 23, 640–650 (2017).
Bergen, P. J. et al. Rational combinations of polymyxins with other antibiotics. Polymyxin antibiotics: From Laboratory Bench to Bedside 1145, 251–288 (2019).
He, J., Abdelraouf, K., Ledesma, K. R., Chow, D. S.-L. & Tam, V. H. Pharmacokinetics and efficacy of liposomal polymyxin B in a murine pneumonia model. Int. J. Antimicrob. Agents 42, 559–564 (2013).
Sans-Serramitjana, E. et al. Killing effect of nanoencapsulated colistin sulfate on Pseudomonas aeruginosa from cystic fibrosis patients. J. Cyst. Fibros 15, 611–618 (2016).
Boge, L. et al. Lipid-based liquid crystals as carriers for antimicrobial peptides: phase behavior and antimicrobial effect. Langmuir 32, 4217–4228 (2016).
Zabara, M. et al. Multifunctional nano‐biointerfaces: cytocompatible antimicrobial nanocarriers from stabilizer‐Free cubosomes. Adv. Funct. Mater. 29, 1904007 (2019).
Lewies, A., Wentzel, J. F., Jordaan, A., Bezuidenhout, C. & Du Plessis, L. H. Interactions of the antimicrobial peptide nisin Z with conventional antibiotics and the use of nanostructured lipid carriers to enhance antimicrobial activity. Int. J. Pharm. 526, 244–253 (2017).
Boge, L. et al. Peptide-loaded cubosomes functioning as an antimicrobial unit against Escherichia coli. ACS Appl. Mater. Interfaces 11, 21314–21322 (2019).
Boge, L. et al. Cubosomes for topical delivery of the antimicrobial peptide LL-37. Eur. J. Pharm. Biopharm. 134, 60–67 (2019).
Meikle, T. G. et al. Incorporation of antimicrobial peptides in nanostructured lipid membrane mimetic bilayer cubosomes. Colloids Surf. B: Biointerfaces 152, 143–151 (2017).
Boge, L. et al. Cubosomes post-loaded with antimicrobial peptides: characterization, bactericidal effect and proteolytic stability. Int. J. Pharm. 526, 400–412 (2017).
Lai, X. et al. Phytantriol-based cubosome formulation as an antimicrobial against lipopolysaccharide-deficient Gram-negative bacteria. ACS Appl. Mater. Interfaces 12, 44485–44498 (2020).
Martinez-Negro, M. et al. Efficient cellular knockdown mediated by siRNA nanovectors of Gemini cationic lipids having delocalizable headgroups and oligo-oxyethylene spacers. ACS Appl. Mater. Interfaces 8, 22113–22126 (2016).
Leal, C., Bouxsein, N. F., Ewert, K. K. & Safinya, C. R. Highly efficient gene silencing activity of siRNA embedded in a nanostructured gyroid cubic lipid matrix. J. Am. Chem. Soc. 132, 16841–16847 (2010).
Kim, H., Song, Z. & Leal, C. Super-swelled lyotropic single crystals. Proc. Natl Acad. Sci. USA 114, 10834–10839 (2017).
Kim, H. & Leal, C. Cuboplexes: topologically active siRNA delivery. ACS Nano 9, 10214–10226 (2015).
Speziale, C., Ghanbari, R. & Mezzenga, R. Rheology of ultraswollen bicontinuous lipidic cubic phases. Langmuir 34, 5052–5059 (2018).
Liu, Z. et al. Cubosome nanoparticles potentiate immune properties of immunostimulants. Int. J. Nanomed. 11, 3571–3583 (2016).
Shi, X. et al. Comparative studies on glycerol monooleate- and phytantriol-based cubosomes containing oridonin in vitro and in vivo. Pharm. Dev. Technol. 22, 322–329 (2017).
Muheem, A., Shakeel, F., Warsi, M. H., Jain, G. K. & Ahmad, F. J. A combinatorial statistical design approach to optimize the nanostructured cubosomal carrier system for oral delivery of ubidecarenone for management of doxorubicin-induced cardiotoxicity: in vitro-in vivo investigations. J. Pharm. Sci. 106, 3050–3065 (2017).
Yang, Z. et al. Development of amphotericin B-loaded cubosomes through the SolEmuls technology for enhancing the oral bioavailability. AAPS PharmSciTech 13, 1483–1491 (2012).
Yeaman, M. R. & Yount, N. Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55, 27–55 (2003).
Velkov, T., Thompson, P. E., Nation, R. L. & Li, J. Structure—activity relationships of polymyxin antibiotics. J. Med. Chem. 53, 1898–1916 (2010).
Clifton, L. A. et al. An accurate in vitro model of the E. coli envelope. Angew. Chem. Int. Ed. Engl. 54, 11952–11955 (2015).
Han, M. L. et al. Polymyxin-induced lipid a deacylation in Pseudomonas aeruginosa perturbs polymyxin penetration and confers high-level resistance. ACS Chem. Biol. 13, 121–130 (2018).
Han, M. L. et al. Investigating the interaction of octapeptin A3 with model bacterial membranes. ACS Infect. Dis. 3, 606–619 (2017).
Shen, H.-H. et al. The interaction of cubosomes with supported phospholipid bilayers using neutron reflectometry and QCM-D. Soft Matter 7, 8041–8049 (2011).
Brandenburg, K. et al. Temperature dependence of the binding of endotoxins to the polycationic peptides polymyxin B and its nonapeptide. Biophys. J. 88, 1845–1858 (2005).
Paracini, N., Clifton, L. A., Skoda, M. W. A. & Lakey, J. H. Liquid crystalline bacterial outer membranes are critical for antibiotic susceptibility. Proc. Natl Acad. Sci. USA 115, E7587–E7594 (2018).
Velkov, T., Thompson, P. E., Nation, R. L. & Li, J. Structure-activity relationships of polymyxin antibiotics. J. Med. Chem. 53, 1898–1916 (2010).
Vaara, M. Agents that increase the permeability of the outer membrane. Microbiol. Mol. Biol. Rev. 56, 395–411 (1992).
Clifton, L. A. et al. Effect of divalent cation removal on the structure of Gram-negative bacterial outer membrane models. Langmuir 31, 404–412 (2015).
Dyett, B. P., Yu, H., Strachan, J., Drummond, C. J. & Conn, C. E. Fusion dynamics of cubosome nanocarriers with model cell membranes. Nat. Commun. 10, 4492 (2019).
Gupta, P., Hung, C. & Perrier, D. Quantitation of the release of doxorubicin from colloidal dosage forms using dynamic dialysis. J. Pharm. Sci. 76, 141–145 (1987).
Zabara, A. et al. Design of ultra-swollen lipidic mesophases for the crystallization of membrane proteins with large extracellular domains. Nat. Commun. 9, 544 (2018).
Astolfi, P. et al. Effects of a cationic surfactant incorporation in phytantriol bulk cubic phases and dispersions loaded with the anticancer drug 5-fluorouracil. J. Mol. Liq. 286, 110954 (2019).
CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 29th Edition. CLSI guideline M100. (Clinical and Laboratory Standards Institute, Wayne, PA, 2019).
CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. 11th Edition CLSI guideline M07. (Clinical and Laboratory Standards Institute, Wayne, PA, 2019).
The European Committee on antimicrobial susceptibility testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 11.0. http://www.eucast.org (2021).
Gales, A. C., Jones, R. N. & Sader, H. S. Contemporary activity of colistin and polymyxin B against a worldwide collection of Gram-negative pathogens: results from the sentry antimicrobial surveillance program (2006–09). J. Antimicrob. Chemother. 66, 2070–2074 (2011).
CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. 6th Editio. CLSI guideline M7-A6. (Clinical and Laboratory Standards Institute, Wayne, PA, 2003).
Han, S. M. et al. Antibacterial activity and antibiotic-enhancing effects of honeybee venom against methicillin-resistant Staphylococcus aureus. Molecules 21, 79 (2016).
James, M. et al. The multipurpose time-of-flight neutron reflectometer “platypus” at Australia’s opal reactor. Nucl. Instrum. Methods Phys. Res. A 632, 112–123 (2011).
Nelson, A. Co-refinement of multiple-contrast neutron/x-ray reflectivity data using MOTOFIT. J. Appl. Crystallogr. 39, 273–276 (2006).
Shen, H. H. et al. Targeted detection of phosphatidylserine in biomimetic membranes and in vitro cell systems using annexin V-containing cubosomes. Biomaterials 34, 8361–8369 (2013).
Reimhult, E., Höök, F. & Kasemo, B. Intact vesicle adsorption and supported biomembrane formation from vesicles in solution: influence of surface chemistry, vesicle size, temperature, and osmotic pressure. Langmuir 19, 1681–1691 (2003).
Rodahl, M., Höök, F., Krozer, A., Brzezinski, P. & Kasemo, B. Quartz crystal microbalance setup for frequency and Q‐factor measurements in gaseous and liquid environments. Rev. Sci. Instrum. 66, 3924–3930 (1995).
Shen, H. H. et al. The influence of dipalmitoyl phosphatidylserine on phase behaviour of and cellular response to lyotropic liquid crystalline dispersions. Biomaterials 31, 9473–9481 (2010).
Acknowledgements
The authors gratefully acknowledge the use of the facilities at the Monash X-ray Platform and Monash Micro Imaging (Monash University, Victoria, Australia). They thank Heidi Yu, Hasini Wickremasinghe and Zhi Ying Kho for their assistance in the in vitro experiment. The Australian Nuclear Science and Technology Organization (ANSTO) is gratefully acknowledged for providing the neutron research facilities (proposal numbers P6945 and P6451). H.-H.S. is an Australian NHMRC Career Development Research Fellow (GNT1106798) and J.L. is an NHMRC Principal Research Fellow (APP1157909). This work was supported by the NHMRC project grant APP1144652.
Author information
Authors and Affiliations
Contributions
X.L. prepared and characterized the structure of samples, carried out PMB-release and microbiological experiments, and performed parts of the confocal and NR experiments, analyzed all the experimental data, wrote the manuscript with contributions from all of the authors. M.L.H. provided the studied strains and subsequent membrane compositions. Y.D. performed the cell viability assay and part of the NR experiment. S.H.C. carried out part of the confocal microscopy and PMB-release experiment. provided experimental and technical support for the SANS experiments. A.P.L.B. and C.-M.W. provided technical support for NR experiments. P.J.B. revised the manuscript. J.-H.J. provided the support for NR data analysis. H.-Y.H. and J.S. revised the manuscript. B.W.M. and J.W. performed the Cryo-TEM test. J.L. provided the foundation support. H.-H.S. conceived the project, provided the foundation support, and modified the structure of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lai, X., Han, ML., Ding, Y. et al. A polytherapy based approach to combat antimicrobial resistance using cubosomes. Nat Commun 13, 343 (2022). https://doi.org/10.1038/s41467-022-28012-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-022-28012-5
This article is cited by
-
Recent advances in therapeutic targets identification and development of treatment strategies towards Pseudomonas aeruginosa infections
BMC Microbiology (2023)
-
Sustained CREB phosphorylation by lipid-peptide liquid crystalline nanoassemblies
Communications Chemistry (2023)
-
Current and evolving knowledge domains of cubosome studies in the new millennium
Journal of Nanoparticle Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.