Abstract
Photoactivation of the plant photoreceptor and thermosensor phytochrome B (PHYB) triggers its condensation into subnuclear membraneless organelles named photobodies (PBs). However, the function of PBs in PHYB signaling remains frustratingly elusive. Here, we found that PHYB recruits PHYTOCHROME-INTERACTING FACTOR 5 (PIF5) to PBs. Surprisingly, PHYB exerts opposing roles in degrading and stabilizing PIF5. Perturbing PB size by overproducing PHYB provoked a biphasic PIF5 response: while a moderate increase in PHYB enhanced PIF5 degradation, further elevating the PHYB level stabilized PIF5 by retaining more of it in enlarged PBs. Conversely, reducing PB size by dim light, which enhanced PB dynamics and nucleoplasmic PHYB and PIF5, switched the balance towards PIF5 degradation. Together, these results reveal that PB formation spatially segregates two antagonistic PHYB signaling actions – PIF5 stabilization in PBs and PIF5 degradation in the surrounding nucleoplasm – which could enable an environmentally sensitive, counterbalancing mechanism to titrate nucleoplasmic PIF5 and environmental responses.
Similar content being viewed by others
Introduction
A universal architectural hallmark of the cell nucleus is various functionally distinct subnuclear membraneless organelles, collectively referred to as nuclear bodies1,2,3. Nuclear bodies act as liquid- or gel-like biomolecular condensates that are commonly thought to be excluded from the surrounding nucleoplasm via concentration-dependent, energetically favorable liquid-liquid phase separation (LLPS)3,4,5. Many nuclear bodies, such as nucleoli, Cajal bodies, nuclear speckles, paraspeckles, histone locus bodies, and promyelocytic leukemia protein (PML) bodies, are well characterized, and they are associated with diverse basic nuclear functions in transcription, splicing, and DNA replication and repair. Accumulating observations have also documented the nonuniform subnuclear distribution of cell signaling molecules to nuclear bodies, in addition to the surrounding nucleoplasm. For instance, the tumor suppressor p53 is recruited to PML bodies6. However, the functional significance of the spatial segregation of signaling molecules into two phase-separated compartments – nuclear bodies and the surrounding nucleoplasm – remains elusive. A better understanding of the function of nuclear bodies in cell signaling requires genetically tractable models directly connecting nuclear body dynamics to cell signaling. Photobodies (PBs) are plant nuclear bodies defined molecularly by the presence of the photoreceptor and thermosensor phytochrome B (PHYB)7,8,9. Environmental light and temperature cues control PHYB activity and, therefore, regulate the assembly/dissolution of PBs and PHYB signaling outputs7,10,11. As such, PBs provide a unique experimental paradigm for interrogating the general principles of nuclear bodies in cell signaling.
PHYs are evolutionarily conserved red (R) and far-red (FR) photoreceptors in plants12. The prototypical plant PHY is a homodimer; each monomer contains an N-terminal photosensory module and a C-terminal output module12,13. PHYs can be photoconverted between two relatively stable forms: the R-light-absorbing inactive Pr form and the FR-light-absorbing active Pfr form12,13. The genome of the reference plant species Arabidopsis thaliana (Arabidopsis) encodes five PHYs, PHYA-E, among which PHYB plays a prominent role14. The photoconversion of PHYB – either photoactivation by R light or photoinhibition by FR light – alters the Pfr/Pr equilibrium of individual PHYB molecules and the steady-state cellular concentration of the active PfrPfr homodimer, thereby allowing plants to monitor changes in light quality, quantity, and periodicity. The amount of active PHYB can also be modulated by ambient temperature through temperature-dependent thermal reversion from Pfr to Pr15, making PHYB a thermosensor in addition to a photoreceptor11,16. By controlling the amount of active PHYB, environmental light and temperature cues regulate all aspects of plant development and growth, including germination, seedling establishment, shade avoidance, floral induction, senescence, and immunity17,18. PHYB signaling is best studied during Arabidopsis seedling establishment, where the perception of environmental cues by PHYB in the epidermal cells of the embryonic leaves (cotyledons) controls the biosynthesis of the mobile growth hormone auxin to regulate seedling morphogenesis, including promoting cotyledon expansion locally, as well as inhibiting the elongation of the embryonic stem (hypocotyl) remotely17,18.
The PHYB-containing subnuclear membraneless organelle was named the “photobody” by Joanne Chory in reference to its dynamic assembly and dissolution in response to R and FR light9. Although “photobody” was also adopted to describe the fascinating blue-light-inducible cryptochrome 2 (CRY2)-containing nuclear bodies19, colocalization studies using BY-2 and Arabidopsis protoplasts suggest that the PHYB-containing PBs and the CRY2-containing condensates may represent distinct nuclear bodies20,21. The regulation of PBs by R/FR light and temperature has been extensively studied in Arabidopsis pavement epidermal cells using fluorescent-protein-tagged PHYB (PHYB-FP). The current model posits that PB formation and maintenance are driven by the condensation of the active PfrPfr form of PHYB. This conclusion is supported by several lines of evidence. First, PB formation and dissolution can be induced by the activation and inactivation of PHYB, respectively. Akira Nagatani, Ferenc Nagy, and Eberhard Schäfer first reported that the photoactivation of PHYB-FP triggers its nuclear accumulation and further compartmentalization into discrete subnuclear speckles (PBs)22,23. Conversely, during the light-to-dark transition or upon an FR treatment where PHYB reverts to the inactive form, PHYB-FP moves from PBs into the nucleoplasm24,25,26,27. Second, the steady-state pattern of PBs is dependent on light intensity and correlates with the amount of active PHYB28. Under strong R light (e.g., 10 μmol m−2 s−1), where each PHYB molecule stays in the Pfr form at least 50% of the time, PHYB-FP assembles into two to ten large PBs of 0.7–2 μm in diameter10,24,28,29. In contrast, under dim R light, where PHYB stays as the Pr form most of the time, PHYB-FP localizes to tens of small PBs of 0.1–0.7 μm in diameter, or it is dispersed evenly in the nucleoplasm10,24,28. Consistent with these observations, a constitutively active phyB mutant YHB, which carries a Y276H mutation in PHYB’s chromophore attachment domain and locks PHYB in an active form, localizes to a few large PBs even in the dark30. In contrast, phyB mutants that destabilize the Pfr form – e.g., removing the disordered N-terminal extension15,31,32 – promote PB dissolution28,31,32,33. Third, PB formation relies on the dimerization of PHYB’s C-terminal module32,34,35. A D1040V mutation in this module, which disrupts PHYB dimerization, abolishes PHYB’s ability to form PBs36. Fourth, PHYB can undergo light-dependent LLPS into biomolecular condensates in mammalian cells37,38. The intramolecular attributes of PHYB required for PB formation in vivo, e.g., the C-terminal module and N-terminal extension, are also essential for PHYB LLPS in this heterologous system37. These observations support the idea that PB formation in vivo may be propelled by the LLPS of active PHYB alone, though PHYB condensation under physiological conditions is likely more complex involving other molecules such as PHOTOPERIODIC CONTROL OF HYPOCOTYL 1 (PCH1)26,39.
PHYB controls diverse developmental responses primarily by regulating a family of basic helix-loop-helix transcription factors called PHYTOCHROME-INTERACTING FACTORs (PIFs)18,40. In general, PIFs are antagonists of PHYB signaling. For instance, the five prominent PIFs – PIF1, PIF3, PIF4, PIF5, and PIF7 – collectively promote hypocotyl elongation18,40. A central mechanism of PHYB signaling is to inhibit the function of PIFs, including promoting their degradation18,40, attenuating their DNA binding41,42, and repressing their transactivation activity43. PIF1, PIF3, PIF4, and PIF5 are rapidly degraded during the dark-to-light transition in a PHYB-dependent manner44,45,46,47,48. PHYB binds directly to the prototypical PIF, PIF3, to promote its phosphorylation by PHOTOREGULATORY PROTEIN KINASEs (PPKs) and subsequent ubiquitin-proteasome-dependent degradation44,49,50,51. Two E3 ubiquitin ligases can ubiquitylate PIF3, including the cullin3-RING E3 ubiquitin ligases (CRL) with LIGHT-RESPONSE BRIC-A-BRAC/TRAMTRACK/BROADs (LRBs) as the substrate recognition subunits (CRL3LRB) and the cullin1-RING E3 with EIN3-BINDING F BOX PROTEINs (EBFs) as the substrate recognition subunits (CRL1EBF)50,51. In addition to regulating PIFs via direct interaction, PHYB also promotes PIF degradation indirectly by inhibiting factors that stabilize PIFs, such as the CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) and SUPPRESSOR OF PHYTOCHROME A-105 proteins (SPAs), which constitute the substrate recognition subunits of the CRL4COP1/SPA E3 ubiquitin ligases52,53,54. Interestingly, although PIF1, PIF3, PIF4, and PIF5 are subject to PHYB-mediated degradation, they exhibit different accumulation patterns. While PIF1 and PIF3 accumulate only in darkness and become undetectable in the light29,36, PIF4 and PIF5 can accumulate significantly under prolonged light treatment36,46,48,55. The accumulation of PIF4 and PIF5 in the light plays an essential role in plants’ responses to warm temperatures during the daytime55,56. However, the mechanism stabilizing PIF4 and PIF5 in the light remains elusive.
PBs are considered to be associated with PHYB signaling7,8. PB formation correlates with PHYB-mediated light responses, such as the inhibition of hypocotyl elongation24,28,29. phyB mutants defective in PB formation are impaired in PHYB signaling28,30,31,32,33,34. Recent proteomics studies revealed that many PHYB signaling components reside in PBs; these components include COP1, SPAs, PPKs, PCH1, PCH1-LIKE (PCHL), and transcription regulators in light and photoperiodic signaling, including PIF4, TANDEM ZINC-FINGER-PLUS 3 (TZP), EARLY FLOWERING 3 (ELF3), EARLY FLOWERING 4 (ELF4), and LUX ARRHYTHMO (LUX), lending further support to the functional role of PBs in PHYB signaling21,26,57. The current data support the model that PBs play a vital role in PIF3 degradation. During the dark-to-light transition, FP-tagged PIF3 localizes to PBs before its degradation44,58. Conversely, PIF3 reaccumulation during the light-to-dark transition correlates with the disappearance of PBs24. Forward genetic screens have identified three PB-deficient mutants, hmr (hemera)29,59,60, rcb (regulator of chloroplast biogenesis)61, and ncp (nuclear control of pep activity)62, in which PHYB-FP fails to form large PBs and PIF3 degradation is blocked in the light. Moreover, overexpressing PHYB’s C-terminal output module alone, which localizes constitutively to PBs, can mediate PIF3 degradation even in darkness36.
Despite the accumulating evidence associating PBs with PIF3 degradation, the precise function of PBs remains frustratingly elusive. We still cannot unequivocally conclude that PBs are the sites of PIF3 degradation, and it is still unknown whether PBs also regulate the stability of other PIFs. A significant challenge in dissecting the function of PBs, and also nuclear bodies in general, has been the difficulty of uncoupling the functional output of the nuclear-body comparment from that of the surrounding nucleoplasm. Because components can diffuse between nuclear bodies and the surrounding nucleoplasm, although manipulating those components may disrupt nuclear body assembly and the functional output associated with the components, such a correlation is usually insufficient to assign the functional defects exclusively to the nuclear body compartment. The previous approach of using loss-of-function mutants in PHYB or PHYB signaling could disrupt the PHYB signaling outputs of either or both PBs and the surrounding nucleoplasm and, therefore, could not uncouple the signaling actions of the two phase-separated compartments. Another hurdle in studying PIF3 degradation was the difficulty in monitoring its PB localization. Because PIF3 does not accumulate to a detectable level in the light, FP-tagged PIF3 can only be observed briefly during the dark-to-light transition44,58.
To circumvent these obstacles to characterizing PBs, we implemented two major strategic changes in the current study. First, we adopted PIF5 as a model because PIF5 accumulates in the light and could potentially allow us to visualize its localization to PBs in light-grown seedlings36,46,48,55, though the PB localization of PIF5 had not been reported previously. Second, instead of disrupting PBs using loss-of-function mutants in PHYB or PHYB signaling, we perturbed the PB size by increasing PHYB abundance. These approaches show that PHYB recruits PIF5 to PBs and, surprisingly, that PHYB exerts two opposing functions in degrading and stabilizing PIF5. Our results reveal that PB formation allows the phase separation and competition of the two antagonistic PHYB signaling actions – PIF5 stabilization in PBs and PIF5 degradation in the nucleoplasm. We propose a PB-enabled counterbalancing mechanism to titrate nucleoplasmic PIF5 and its signaling outputs.
Results
Increasing PHYB production alters PB size and dynamics
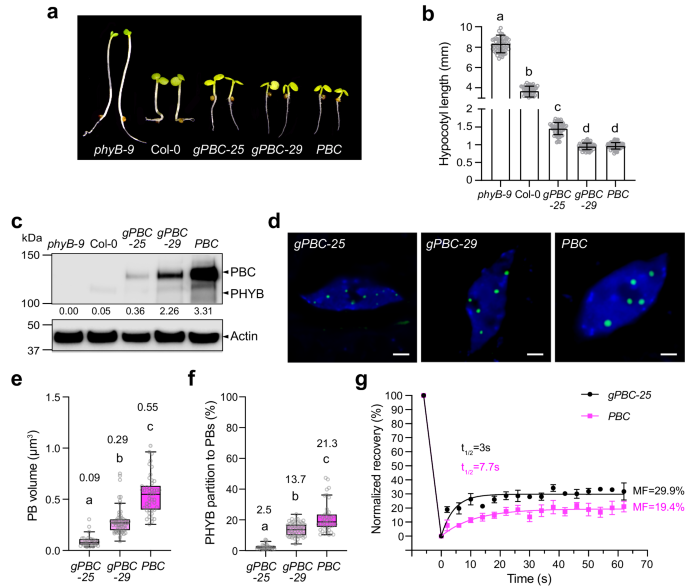
Instead of interrogating the function of PBs in loss-of-function phyB or PHYB signaling mutants, we explored an alternative approach to perturb PBs by increasing PHYB abundance. The current hypothesis is that PBs form via the LLPS of PHYB37. Based on the theory of the LLPS, the LLPS of PHYB occurs when PHYB accumulates at a critical concentration. The PHYB LLPS model predicts that the overproduction of PHYB above the critical concentration is expected to result only in the growth of PBs without changing the concentration of PHYB in either the PB or the surrounding nucleoplasmic compartment4,63. Therefore, if the model were correct, increasing the PHYB level should only enhance the signaling output from PBs while leaving the functional output of the nucleoplasmic PHYB unchanged or less affected. To test this hypothesis, we collected Arabidopsis lines expressing various amounts of PHYB. We previously reported the PBC line, which overexpresses CFP-tagged PHYB (PHYB-CFP) in the phyB-9 background32. PBC exhibits a short hypocotyl phenotype, as it overexpresses PHYB-CFP at a level 65-fold that of the endogenous PHYB (Fig. 1a–c)32. To create lines expressing intermediate levels of PHYB between PBC and Col-0, we generated new transgenic lines in the phyB-9 background carrying PHYB genomic DNA with a CFP sequence inserted immediately before the PHYB stop codon. We named them gPBC (genomic PHYB-CFP) lines. Two single-insertion gPBC lines, gPBC-25 and gPBC-29, were selected for further analysis. The steady-state levels of PHYB-CFP in gPBC-25 and gPBC-29 were about 7- and 40-fold that of the endogenous PHYB level in Col-0, respectively (Fig. 1a–c). Both gPBC lines rescued the long-hypocotyl phenotype of phyB-9, indicating that PHYB-CFP was functional (Fig. 1a, b). Corroborating the notion that the PHYB response correlates with the PHYB level, the hypocotyl length of gPBC-25 was in between that of Col-0 and PBC, whereas the hypocotyl length of gPBC-29 was similar to that of PBC (Fig. 1a, b). With the two new gPBC lines, we had a panel of five lines – phyB-9, Col-0, gPBC-25, gPBC-29, and PBC – with PHYB levels ranging from zero to 65-fold that of the wild-type level (Fig. 1a–c).
a Images of 4-d-old phyB-9, Col-0, gPBC-25, gPBC-29 and PBC seedlings grown under 10 μmol m−2 s−1 R light at 21 oC. b Hypocotyl length measurements of the seedlings shown in (a). Error bars represent the s.d. (n > 50 seedlings); the centers of the error bars indicate the mean. Different letters denote statistically significant differences in hypocotyl length (ANOVA, Tukey’s HSD, multiplicity adjusted p ≤ 0.05). c Immunoblots showing the PHYB levels in the seedlings described in (a). Actin was used as a loading control. The relative endogenous PHYB and PHYB-CFP levels normalized to the corresponding levels of actin are shown. The immunoblot experiments were independently repeated three times with similar results. d Confocal images showing representative PHYB-CFP PB patterns in cotyledon pavement epidermal cells from 4-d-old gPBC-25, gPBC-29 and PBC seedlings grown under 10 μmol m−2 s−1 R light at 21 oC. Scale bars are equal to 2 µm. e Quantification of the PB volume in cotyledon epidermal cells from gPBC-25, gPBC-29 and PBC as shown in (d). Different letters denote statistically significant differences in volume (ANOVA, Tukey’s HSD, multiplicity adjusted p < 0.05). f Quantification of the percentage of PHYB partitioned to PBs based on the total PHYB signal within the nuclei of gPBC-25, gPBC-29 and PBC as shown in (d). Box and whisker plots showing the percent of PHYB partitioned to PBs per nucleus. Different letters denote statistically significant differences in percentage (ANOVA, Tukey’s HSD, multiplicity adjusted p < 0.05). For (e, f), the numbers indicate the mean values. In the box and whisker plots, the boxes represent the 25th to 75th percentiles, and the bars are equal to the median. g Results of FRAP experiments showing normalized fluorescence recovery plotted over time for PHYB-CFP in PBs of pavement cell nuclei from 4-d-old gPBC-25 (black) and PBC (magenta) seedlings grown under 50 μmol m−2 s−1 R light. Values represent the mean, and error bars represent the s.e. of the mean of three biological replicates. The solid lines represent an exponential fit of the data. MF: mobile fraction; t1/2 represents the half-time of fluorescence recovery. The source data underlying the hypocotyl measurements in (b), the immunoblots in (c), and the PB characterization in (e–g) are provided in the Source Data file.
To test whether increasing the PHYB level enhances the size of PBs, we measured the volumes of PHYB-CFP PBs in gPBC-25, gPBC-29, and PBC. Indeed, the PB size increased with the PHYB-CFP level (Fig. 1d, e). PBC had the largest PBs, which were, on average, more than five-fold larger than those in gPBC-25. The average PB volume of gPBC-29 was three times larger than that of gPBC-25 (Fig. 1e). The fraction of PHYB-CFP localized to PBs also increased with the PHYB-CFP level (Fig. 1f). These results support the predictions of the PHYB LLPS model that increasing PHYB abundance enlarges PBs. However, these data did not assess whether the concentration of PHYB-CFP remained the same in the PBs and the surrounding nucleoplasm. We used fluorescence recovery after photobleaching (FRAP) to evaluate the exchange of PHYB-CFP molecules between PBs and the surrounding nucleoplasm. To our surprise, the PBs from gPBC-25 and PBC exhibited significant differences in the dynamics of PHYB-CFP. The fluorescence recovery in gPBC-25 was only 29.9%, suggesting that the majority of PHYB-CFP molecules were not mobile (Fig. 1g). The percentage of fluorescence recovery in PBC was further reduced to 19.4%, indicating that increasing PHYB-CFP abundance decreased the mobile fraction of PHYB-CFP in PBs, likely due to a transition to a gel-like state (Fig. 1g). Supporting this idea, PHYB-CFP in the PBs from PBC showed a decrease in the fluorescence recovery kinetics compared with gPBC-25, indicating a reduction in the diffusion rate of PHYB-CFP in PBC (Fig. 1g). Together, these results demonstrate that increasing PHYB abundance enhances PB size by recruiting a larger fraction of PHYB to the PB compartment and also induces a transition of PBs to a gel-like state that could retain PHYB for a longer time in PBs. Thus, this panel of lines with various PB sizes and PHYB-CFP dynamics provides an opportunity to interrogate the function of PBs in PIF degradation.
PIF5 is a short-lived protein localized in PBs
To investigate the function of PBs in PIF regulation, we turned to PIF5 as a model because, despite PHYB-mediated PIF5 degradation45,46,48, PIF5 can accumulate in the light36,46,48,55,64, which could potentially allow us to monitor its subnuclear localization in the light. We generated transgenic lines expressing PIF5 fused with Myc and mCherry to the N-terminus (mCherry-PIF5) under the native PIF5 promoter in the null pif5-3 mutant background. Two independent transgenic lines with a single insertion of the transgene, named mCherry-PIF5/pif5-3 #8 and #9, were selected for further analysis. The pif5-3 mutant exhibited a short hypocotyl phenotype at 16 oC. Because PIF5 is required for the warm temperature-induced hypocotyl elongation, the defect of pif5-3 in hypocotyl growth became more pronounced at 27 oC (Fig. 2a, b)55. The mCherry-PIF5/pif5-3 lines rescued the hypocotyl-growth defects of pif5-3 at both temperatures (Fig. 2a, b), indicating that mCherry-PIF5 was functional. Similar to endogenous PIF5, mCherry-PIF5 accumulated in the light and could be detected by immunoblotting using anti-PIF5 antibodies (Fig. 2c). Despite being controlled by the native PIF5 promoter, the steady-state levels of mCherry-PIF5 in the transgenic lines were higher than that of endogenous PIF5 in Col-0. The mCherry-PIF5 level in line #8 was more than five-fold that of line #9 and more than ten-fold that of endogenous PIF5 in Col-0 (Fig. 2c). However, to our surprise, no mCherry signal could be detected by confocal microscopy in either of the mCherry-PIF5/pif5-3 transgenic lines. We reasoned that this discrepancy might be attributable to a faster turnover rate of mCherry-PIF5 compared with the maturation time required for newly synthesized mCherry to become fluorescent. The reported maturation time of mCherry is more than 60 min65. If the half-life of mCherry-PIF5 is significantly shorter than 60 min, the newly synthesized mCherry-PIF5 would have been degraded before becoming fluorescent. If this were the case, the non-fluorescent mCherry-PIF5 protein should be detectable by immunolocalization. To test the hypothesis, we performed immunofluorescence staining using anti-Myc antibodies. Indeed, Myc-tagged mCherry-PIF5 could be detected by immunostaining. More interestingly, mCherry-PIF5 was localized to discrete punctate structures at both 16 oC and 27 oC (Fig. 2d). Simultaneously labeling endogenous PHYB using anti-PHYB antibodies demonstrated that mCherry-PIF5 colocalized with PHYB in PBs (Fig. 2d, e). Thus, these results indicate that mCherry-PIF5 is a short-lived protein that colocalizes with PHYB in PBs under the physiological PHYB concentration.
a Images of 4-d-old seedlings of Col-0, pif5-3 and two mCherry-PIF5/pif5-3 (#8 and #9) lines grown under 50 μmol m−2 s−1 R light at 16 oC or 27 oC. b Hypocotyl length measurements of the seedlings described in a. The open and gray bars represent hypocotyl length measurements at 16 oC and 27 oC, respectively. Error bars for the hypocotyl measurements represent the s.d. (n = 90 seedlings). Lowercase and uppercase letters denote statistically significant differences in hypocotyl lengths at 16 oC or 27 oC, respectively (ANOVA, Tukey’s HSD, multiplicity adjusted p < 0.05, n = 90 seedlings). The black number above the Col-0 data represents the percent increase in hypocotyl length at 27 oC compared to 16 oC (mean ± s.d., n = 90 seedlings). The pink bars show the relative response, defined as the hypocotyl response to 27 oC of a mutant or transgenic line relative to that of Col-0 (set at 100%). Error bars for the relative responses represent the s.e. of three biological replicates. The centers of all error bars indicate the mean. Pink numbers show the mean ± s.e. of the relative responses. Different pink letters denote statistically significant differences in the relative responses (ANOVA, Tukey’s HSD, multiplicity adjusted p < 0.05, n = 3 biological replicates). c Immunoblots showing the steady-state levels of PIF5 and mCherry-PIF5 in 4-d-old Col-0, pif5-3, and the mCherry-PIF5 lines (#8, #9) grown under 50 μmol m−2 s−1 R light at 16 oC or 27 oC. PIF5 and mCherry-PIF5 were detected using anti-PIF5 antibodies. Actin was used as a loading control. The relative levels of PIF5 or mCherry-PIF5 normalized to actin are shown. The asterisk indicates a nonspecific band. d Confocal images showing the colocalization of mCherry-PIF5 and PHYB in PBs in cotyledon pavement epidermal nuclei from 4-d-old mCherry-PIF5 line #8 seedlings grown under 50 μmol m−2 s−1 R light at 16 oC or 27 oC. PHYB (green) and Myc-tagged mCherry-PIF5 (red) were labeled via immunofluorescence staining using anti-PHYB and anti-Myc antibodies, respectively. Nuclei were stained with DAPI (blue). Scale bars are equal to 2 µm. e Quantification of the colocalization of mCherry-PIF5 foci with PHYB-CFP PBs in the experiments described in (d). Error bars represent the s.e. of at least three biological replicates. The source data underlying the hypocotyl measurements in (b), the immunoblots in (c), and the PB characterization in (e) are provided in the Source Data file.
PHYB signaling both degrades and stabilizes PIF5
The fact that mCherry-PIF5 was short-lived and localized in PBs might suggest that mCherry-PIF5 degradation occurs in PBs. However, it was equally possible that mCherry-PIF5 was degraded in the nucleoplasm and that because mCherry-PIF5 was exchanged between the nucleoplasm and PBs, both the nucleoplasmic and PB pools of mCherry-PIF5 were short-lived. One way to distinguish between these two possibilities would be to test whether perturbing PBs alters PIF5 degradation. If PIF5 were degraded in PBs, increasing the size of PBs should recruit more PIF5 to PBs and accelerate PIF5 degradation. To test the hypothesis, we examined the endogenous levels of PIF5 in the panel of lines expressing various amounts of PHYB (Fig. 1a–c). Surprisingly, PIF5 showed a biphasic response to increases in PHYB abundance. In the range of zero to moderate levels of PHYB in phyB-9, Col-0, and gPBC-25, the steady-state level of PIF5 decreased with the increases in PHYB (Fig. 3a–c), supporting the current model that PHYB promotes PIF5 degradation in the light. However, further increasing PHYB abundance in gPBC-29 and PBC unexpectedly enhanced PIF5 accumulation (Fig. 3a, b). Despite the changes in the PHYB level, the transcript levels of PIF5 remained the same in phyB-9, Col-0, gPBC-25, gPBC-29, and PBC (Fig. 3c). Therefore, the changes in the protein level of PIF5 must be due to either PIF5 translation or degradation. We next examined the degradation kinetics of PIF5 in Col-0, gPBC-25, and PBC by treating the cell lysates with the translation inhibitor cycloheximide and then monitoring the disappearance of PIF5 over time. PIF5 was degraded much faster in gPBC-25 than in Col-0 (Fig. 3d, e). Surprisingly, however, PIF5 degradation was attenuated dramatically in PBC. Thus, the accumulation of PIF5 in PBC and gPBC-29 was most likely due to the PHYB-mediated stabilization of PIF5. These results suggest that PHYB signaling exerts opposing roles in both degrading and stabilizing PIF5, the balance of which can be adjusted by altering the PHYB level. Because the size of PBs increased significantly from gPBC-25 to PBC (Fig. 1d, e), these results raised the hypothesis that while PHYB promotes PIF5 degradation in the nucleoplasm, it stabilizes PIF5 in PBs. As such, increasing the PB size in PBC could switch the balance towards PIF5 stabilization by recruiting more PIF5 to PBs.
a Immunoblots showing PHYB and PIF5 levels in 4-d-old phyB-9, Col-0, gPBC-25, gPBC-29 and PBC seedlings grown under 10 μmol m−2 s−1 R light at 21 oC. 4-d-old dark-grown Col-0 and R-light-grown pifq were positive and negative controls for PIF5, respectively. The immunoblot experiments were independently repeated three times with similar results. b Quantification of the PHYB and PIF5 levels shown in (a). Error bars represent the s.d. of three independent replicates. The centers of the error bars indicate the mean values. Different lowercase and uppercase letters denote statistically significant differences in the abundance of PHYB and PIF5, respectively (ANOVA, Tukey’s HSD, multiplicity adjusted p < 0.05, n = 3 biological replicates). c Quantitative real-time PCR (qRT-PCR) analysis of the transcript levels of PHYB and PIF5 in seedlings shown in (a). Different lowercase and uppercase letters denote statistically significant differences in PHYB and PIF5 transcripts, respectively (ANOVA, Tukey’s HSD, multiplicity adjusted p < 0.05, n = 3 biological replicates). d Immunoblots showing the degradation kinetics of PIF5 in Col-0, gPBC-25, and PBC. 4-d-old Col-0, gPBC-25, and PBC seedlings were incubated with cycloheximide and collected at the indicated time points. Total proteins were extracted and subjected to immunoblotting analysis using anti-PIF5 antibodies. For (a, d), actin was used as a loading control. The asterisk indicates a nonspecific band. The relative PHYB or PIF5 levels normalized to the corresponding actin levels are shown. e Quantification of the relative PIF5 protein levels shown in (d). Error bars represent the s.d. of three independent replicates. The source data underlying the immunoblots in (a, b, d, e), and the qRT-PCR analysis in (c) are provided in the Source Data file.
PHYB recruits and stabilizes PIF5 in PBs
To further examine the role of PBs in stabilizing PIF5, we reasoned that if PIF5 were stabilized due to its enhanced recruitment to the enlarged PBs in PBC, we might be able to observe the accumulation of stabilized or longer-lived mCherry-PIF5 proteins in PBs in PBC via confocal microscopy. To that end, we generated transgenic lines expressing mCherry-PIF5 tagged with human influenza hemagglutinin (HA) (HA-mCherry-PIF5) under the constitutive UBIQUITIN 10 promoter in PBC (mCherry-PIF5/PBC). We selected two mCherry-PIF5/PBC lines (#1 and #9) for further analysis. The mCherry-PIF5/PBC lines were taller than PBC at 16 oC and 27 oC, suggesting that the HA-tagged mCherry-PIF5 was functional (Fig. 4a, b). As endogenous PIF5, mCherry-PIF5 could accumulate in both mCherry-PIF5/PBC lines in the light at 16 oC and 27 oC (Fig. 4c). Similar to the mCherry-PIF5/pif5-3 lines (Fig. 3c), the mCherry-PIF5 levels in the mCherry-PIF5/PBC lines were more than ten-fold greater than that of the endogenous PIF5 in Col-0 (Fig. 4c). However, unlike the mCherry-PIF5/pif5-3 lines, we could observe the fluorescent signal of mCherry-PIF5 in the mCherry-PIF5/PBC lines using confocal microscopy (Fig. 4d). mCherry-PIF5 was also colocalized with PHYB-CFP in PBs in the mCherry-PIF5/PBC lines at both 16 oC and 27 oC in the light (Fig. 4d, e). In dark-grown mCherry-PIF5/PBC seedlings where there were no PHYB-CFP PBs, mCherry-PIF5 was evenly dispersed in the nucleoplasm (Fig. 4f), implying that mCherry-PIF5 was recruited to PBs in the light by PHYB. The fact that mCherry-PIF5 became detectable by confocal microscopy suggested that the half-life of the mCherry-PIF5 in mCherry-PIF5/PBC was significantly longer than that in mCherry-PIF5/pif5-3. Supporting this conclusion, mCherry-PIF5 accumulated to higher levels in mCherry-PIF5/PBC than mCherry-PIF5/pif5-3 (Figs. 3a, b and 4c). Moreover, the degradation kinetics of mCherry-PIF5 in mCherry-PIF5/PBC became slower than that in mCherry-PIF5/pif5-3 (Fig. 4g, h). It is important to note that the degradation kinetics of mCherry-PIF5 (Fig. 4g, h) was considerably faster than that of endogenous PIF5 (Fig. 3d, e)48, this could be attributed to either the mCherry tag or the overexpression of mCherry-PIF5 and the resulted change in mCherry-PIF5’s PB/nucleoplasm partitioning. Together, these results support the model that PHYB recruits and stabilizes PIF5 in PBs and that the action of PHYB in PIF5 stabilization is enhanced in PBC by retaining more PIF5 in the enlarged PBs.
a Images of 4-d-old PBC and mCherry-PIF5/PBC (#1 and #9) seedlings grown under 10 μmol m−2 s−1 R light at either 16 oC or 27 oC. b Hypocotyl length measurements of the seedings described in (a). Error bars represent the s.d., and the centers represent the mean (n ≥ 32 seedlings). Samples labeled with letters exhibited statistically significant differences in hypocotyl length (ANOVA, Tukey’s HSD, multiplicity adjusted p < 0.05). c Immunoblots showing the steady-state levels of PIF5 and mCherry-PIF5 in 4-d-old pif5-3, Col-0, PBC, and mCherry-PIF5/PBC (#1, #9) seedlings grown under 10 μmol m−2 s−1 R light at either 16 oC or 27 oC. d Confocal images showing the colocalization of mCherry-PIF5 with PHYB-CFP in PBs in cotyledon pavement cells from 4-d-old mCherry-PIF5/PBC #9 seedlings grown under 10 μmol m−2 s−1 R light at either 16 oC or 27 oC. Scale bars are equal to 2 µm. e Quantification of the colocalization of mCherry-PIF5 foci with PHYB-CFP PBs in the experiments described in (d). Error bars represent the s.e. (n = 9 biological replicates for the 16 oC samples, n = 7 biological replicates for the 27 oC samples), and the centers represent the mean. f Fluorescence microscope images showing the localization of PHYB-CFP and mCherry-PIF5 in hypocotyl epidermal cells from 4-d-old dark-grown mCherry-PIF5/PBC #9 seedings. The DIC image shows the nucleus. Scale bars are equal to 5 µm. g Immunoblots showing the degradation kinetics of Myc-tagged mCherry-PIF5 in mCherry-PIF5/pif5-3 and HA-tagged mCherry-PIF5 in mCherry-PIF5/PBC. 4-d-old seedlings of mCherry-PIF5/pif5-3 #8 and mCherry-PIF5/PBC #9 were incubated with cycloheximide and collected at the indicated time points. For (c, g), PIF5 and mCherry-PIF5 were detected using anti-PIF5 antibodies. Actin was used as a loading control. The asterisk indicates a nonspecific band. The relative levels of PIF5 and mCherry-PIF5 normalized to the corresponding actin levels are shown below each lane. h Quantification of the relative mCherry-PIF5 levels shown in (g). Error bars represent the s.d. of four independent replicates. The source data underlying the hypocotyl measurements in (b), the immunoblots in (c, g, h), and the quantification of the colocalization of mCherry-PIF5 and PHYB-CFP in e are provided in the Source Data file.
Reducing PB size accelerates PIF5 degradation
If the formation of PBs phase separates the opposing PHYB signaling actions of PIF5 degradation and stabilization, altering PB size and, therefore, the PB/nucleoplasm partitioning of PHYB and PIF5 should be able to shift the balance of the two antagonistic PHYB actions and result in a change in the level of PIF5 and its signaling output. For example, if the above model is correct, reducing PB size in mCherry-PIF5/PBC under dimmer light should increase the nucleoplasmic fractions of PHYB and PIF5 and rebalance PHYB signaling from stabilizing PIF5 to degrading PIF5. To test this hypothesis, we compared the size and dynamics of PBs and PIF5 degradation in mCherry-PIF5/PBC #9 grown under 10 μmol m−2 s−1 or 0.5 μmol m−2 s−1 R light. As expected, the dimmer R light reduced PHYB activity and promoted hypocotyl elongation in Col-0 (Fig. 5a, b). The mCherry-PIF5/PBC seedlings grown under 0.5 μmol m−2 s−1 R light were also significantly taller than the mCherry-PIF5/PBC seedlings grown under 10 μmol m−2 s−1 and PBC seedlings grown 0.5 μmol m−2 s−1 R light (Fig. 5a, b), which suggests that mCherry-PIF5 in mCherry-PIF5/PBC was more abundant and/or more active under the dim light. As expected, PHYB-CFP PBs became significantly smaller under the dim light (Fig. 5c, d)28. Interestingly, mCherry-PIF5 was detectable using confocal microscopy in only 75.3% of PBs in the dim light (Fig. 5e) as opposed to in 100% PBs under strong R light (Fig. 4e), suggesting that mCherry-PIF5 was less stable in dim light. Supporting this conclusion, the steady-state levels of both mCherry-PIF5 and endogenous PIF5 decreased dramatically in mCherry-PIF5/PBC under 0.5 μmol m−2 s−1 R light compared to 10 μmol m−2 s−1 R light (Fig. 5f). The degradation kinetics of mCherry-PIF5 were dramatically increased in mCherry-PIF5/PBC under dim R light compared to that under strong R light (Fig. 5g). PHYB-CFP became more dynamic in the small PBs under 0.5 μmol m−2 s−1 R light, suggesting that dimmer light promoted a gel-to-liquid transition (Fig. 5h). Intriguingly, the dynamics of mCherry-PIF5 stayed the same between high and low light conditions (Fig. 5i). These results suggest that although the large and small PBs might have different PIF5-binding capacities, their PIF5 binding affinities were similar. Therefore, reducing the PB size promoted PIF5 degradation likely by enhancing the partitioning of PHYB and PIF5 to the nucleoplasm compartment. Together, these results provide further evidence supporting our conclusion that recruiting PIF5 to PBs stabilizes PIF5 by preventing its degradation in the surrounding nucleoplasm.
a Images of 4-d-old Col-0, PBC and mCherry-PIF5/PBC #9 seedlings grown under either 10 μmol m−2 s−1 or 0.5 μmol m−2 s−1 R light (R-10 and R-0.5, respectively). b Hypocotyl length measurement of the seedlings described in (a). Error bars represent the s.d., and the centers represent the mean (n = 90 seedlings). Samples labeled with different letters exhibited statistically significant differences in hypocotyl length (two-tailed Student’s t-test, p < 0.05). c Confocal images showing the PB patterns in cotyledon pavement epidermal cells from the mCherry-PIF5/PBC seedlings described in (a). Scale bars equal 2 µm. d Quantification of the sizes of the PBs described in (c). Error bars represent the s.d., and the center represents the mean (n = 53 PBs for the R-10 samples, n = 72 for the R-0.5 samples). Different letters denote statistically significant differences in PB size (two-tailed Student’s t-test, p < 0.05). e Quantification of the percentage PHYB-CFP PBs with detectable mCherry-PIF5 fluorescence in mCherry-PIF5/PBC seedlings grown under 0.5 μmol m−2 s−1 R light. The error bar represents the s.d., and the center represents the mean (n = 30). f Immunoblots of PHYB-CFP, endogenous PIF5 and mCherry-PIF5 in 4-d-old PBC and mCherry-PIF5/PBC #9 seedlings grown under either 10 μmol m−2 s−1 or 0.5 μmol m−2 s−1 R light. PHYB-CFP was detected using anti-PHYB antibodies, and mCherry-PIF5 and PIF5 were detected using anti-PIF5 antibodies. Actin was used as a control. The relative levels of PHYB-CFP, endogenous PIF5, and mCherry-PIF5, normalized to the corresponding actin levels, are shown under each lane. The asterisk indicates a nonspecific band. g Immunoblots showing the degradation kinetics of mCherry-PIF5 in 4-d-old mCherry-PIF5/PBC #9 seedlings grown under either 10 μmol m−2 s−1 or 0.5 μmol m−2 s−1 R light. Seedlings were incubated with cycloheximide and collected at the indicated time points. Total proteins were extracted and subjected to immunoblotting analysis using anti-PIF5 antibodies. Actin was used as a loading control. The relative mCherry-PIF5 levels normalized to the corresponding levels of actin are shown. h Results of FRAP experiments showing normalized fluorescence recovery plotted over time for PHYB-CFP in the mCherry-PIF5/PBC #9 seedlings described in (a). i Results of FRAP experiments showing normalized fluorescence recovery plotted over time for mCherry-PIF5 in the mCherry-PIF5/PBC #9 seedlings described in (a). For (h, i), values represent the mean, and error bars represent the s.e. of at least three biological replicates. Solid lines represent the exponential fit of the data. MF: mobile fraction. t1/2 represents the half-time of fluorescence recovery. The source data underlying the hypocotyl measurements in (b), the PB characterization in (d, e), the immunoblots in (f, g), and the FRAP results in (h, i) are provided in the Source Data file.
Discussion
Reports published more than 20 years ago described how light triggers the localization of photoactivated PHYB to discrete PBs, creating two spatially separated pools of PHYB – a concentrated pool in PBs and a dilute pool in the surrounding nucleoplasm – and that PB size depends on the amount of active PHYB and could be directly regulated by light intensity and quality (i.e., the R/FR ratio)22,23,28. However, the functional significance of this spatial segregation of active PHYB remained frustratingly elusive. Here, we show that PHYB recruits PIF5 as a client to PBs and, surprisingly, that PHYB exerts two opposing functions in degrading and stabilizing PIF5. Unlike previous studies on PBs, we perturbed PBs by increasing PHYB abundance. This approach allowed us to uncouple the function of PHYB in PBs from that in the surrounding nucleoplasm. Our results support the model that the condensation of PHYB phase separates the opposing PHYB signaling actions of PIF5 degradation and stabilization into two subnuclear compartments: a PIF5-stabilizing environment in PBs and a PIF5-degrading environment in the surrounding nucleoplasm (Fig. 6a). As such, PB dynamics may regulate the PB/nucleoplasmic partitioning of PHYB and PIF5 and thus enable an environmentally sensitive counterbalancing mechanism for titrating the nucleoplasmic concentration of PIF5 and PHYB signaling outputs (Fig. 6b). Because control of the stability of PHYB-associated signaling components is a major mechanism of light signaling9,18,40, this PB-dependent counterbalancing mechanism for PIF5 regulation provides a framework for assessing the function of PBs in regulating other PHYB-interacting signaling molecules in light and temperature signaling. We propose that this PB function represents a general function of biomolecular condensates that allows distinct variations of a cellular process or signaling pathway to coexist and interact to generate dynamically adjustable integrated outputs within a single subcellular space.
a Schematic illustration of the function of PBs in segregating the opposing PHYB signaling actions of PIF5 degradation and stabilization. The condensation of PHYB creates a PIF5-stabilizing environment in PBs to counteract PIF5 degradation in the surrounding nucleoplasm. PHYB promotes PIF5 degradation in the nucleoplasm by triggering its phosphorylation and subsequent ubiquitylation by the CRL4COP1/SPA E3 ubiquitin ligases45,46,48. PHYB serves as a scaffold component to recruit PIF5 as a client to PBs via direct interaction. PHYB stabilizes PIF5 by selectively recruiting COP1 and SPAs, but not the CRL4 core, to PBs21,26,71. The high concentration of PHYB in PBs disrupts the COP1-SPA complex via distinct interactions of COP1 with PHYB’s N-terminal photosensory module and SPAs with PHYB’s C-terminal output module52,53,54,68,70. b Model for a PB-enabled counterbalancing mechanism to titrate nucleoplasmic PIF5 and environmental responses. Changes in the intensity and composition of light (and also in temperature) directly control the amount of active PHYB and thus the size of PBs to regulate the PB-to-nucleoplasm partitioning of PHYB, PIF5, and other PB constituents, thereby titrating the nucleoplasmic PIF5 and its signaling output. Strong R light increases the amount of active PHYB and enlarges PBs, thereby sequestering a greater fraction of PHYB and PIF5 in PBs, which stabilizes PIF5 and simultaneously reduces the nucleoplasmic PIF5 and its signaling output. This mechanism allows hypocotyl to grow slowly in the light. Dim R light or shade reduces the amount of active PHYB and PB size, thereby enhancing the nucleoplasmic fraction of PHYB and PIF5, simultaneously promoting PIF5 degradation and its transcriptional output. The latter mechanism accelerates hypocotyl elongation.
Our results indicate that PHYB recruits PIF5 to PBs for PIF5 stabilization. We demonstrated for the first time that PIF5 localizes to PBs. This conclusion is supported by the colocalization of mCherry-PIF5 with endogenous PHYB in PBs in mCherry-PIF5/pif5-3 and with PHYB-CFP PBs in mCherry-PIF5/PBC (Figs. 2d, e, 4d, e). We showed that the slow maturation time of FPs poses a major obstacle in observing FP-tagged PIFs in live cells. The short-lived mCherry-PIF5 in the mCherry-PIF5/pif5-3 lines could only be detected using immunofluorescence staining, not via confocal microscopy (Fig. 2d). This technical hurdle might explain the surprisingly low number of detailed investigations into the subcellular localization of FP-tagged PIFs compared with the overwhelming number of reports on PIFs’ functions. Although the slow maturation of mCherry hindered our ability to observe mCherry-PIF5, we serendipitously found that the detectability of mCherry fluorescence could be used as an internal reporter to assess the stability or half-life of mCherry-PIF5 in live cells. When PIF5 degradation was attenuated in mCherry-PIF5/PBC (Fig. 4g), the fluorescence of mCherry-PIF5 became detectable using confocal microscopy (Fig. 4d). The fact that longer-lived mCherry-PIF5 in mCherry-PIF5/PBC was colocalized with PHYB-CFP in PBs provides direct evidence demonstrating that mCherry-PIF5 was retained and stabilized in PBs (Fig. 4d, e). Interestingly, mCherry-PIF5 in mCherry-PIF5/PBC localized to PBs only in the light but not in darkness (Fig. 4d, f), supporting the idea that PIF5 was recruited to PBs by PHYB, likely via direct interaction45. This conclusion corroborates the proteomics results that PBs comprise PHYB and its primary and secondary interacting signaling components21,26. PIF5 was not identified in the reported proteomics analysis of PB components, which may be due to the timing and conditions used in those studies, as the expression of PIF5 was controlled by the circadian clock66. One concept of biomolecular condensates is the concept of scaffolds and clients67. Scaffold molecules are considered the drivers of phase separation, whereas molecules recruited into biomolecular condensates formed by scaffolds are called clients67. The current data suggest that PHYB is a scaffold component, whereas PIF5 is a client component recruited to PBs, likely via direct interactions with PHYB and other PB components (Fig. 6a). Supporting this model, altering PB size by manipulating the concentration of active PHYB changed the dynamics of the scaffold PHYB but not that of PIF5 (Fig. 5h, i), suggesting that changing PB size altered only PB’s binding capacity but not affinity to PIF5. Together, our results reveal that in addition to promoting PIF5 degradation, PHYB stabilizes PIF5 by recruiting PIF5 to PBs, providing a mechanism that allows PIF5 to accumulate in the light.
PHYB-mediated degradation of PIFs has been invoked as one the most important mechanisms of light signaling18,40. PIF5 is relatively stable in dark-grown seedlings, and upon exposure to light, PHYA and PHYB induce the rapid phosphorylation and degradation of PIF545,46,48. However, the site of PIF5 degradation has yet to be previously studied. Our results support the model that the PHYB condensation phase separates the PIF5-stabilizing environment in PBs from the PIF5-degrading environment in the surrounding nucleoplasm (Fig. 6a). The mechanism that distinguishes the PIF5-stabilizing environment in PBs and the PIF5-degrading environment in the surrounding nucleoplasm requires further investigation. Previous studies have suggested that PIF5 degradation is mediated by the CRL4COP1/SPA E3 ubiquitin ligase48. PHYB interacts directly with COP1 and SPAs via different domains: while the N-terminal photosensory module interacts with COP168, the C-terminal output module confers a strong interaction with SPAs52,53,69. As such, PHYB inhibits the activity of CRL4COP1/SPA by blocking the COP1-SPA interaction52,53,54,68,70. Both COP1 and SPAs have been identified as PB components21,26. It is likely that the high concentration of PHYB in PBs facilitates the dissociation of COP1-SPA complexes (Fig. 6a). In addition, cullin4 was not identified as a PB constituent26,57 and, instead, was shown to disperse evenly in the nucleoplasm71. Therefore, it is possible that the functional CRL4COP1/SPA complex could be assembled only in the nucleoplasm, providing another possible mechanism for the distinct functions between PBs and the surrounding nucleoplasm (Fig. 6a). In this model, the dynamic changes of PBs would also regulate the amount of functional CRL4COP1/SPA complexes in the nucleoplasm.
Our results suggest that PHYB condensation may enable an environmentally sensitive counterbalancing mechanism to titrate environmental responses (Fig. 6b). Changes in light intensity and composition (and also temperature) directly control the amount of active PHYB and the size of PBs, which could regulate the PB-to-nucleoplasmic partitioning of PHYB, PIF5, and other PB constituents, thereby titrating the nucleoplasmic PIF5 and its signaling output (Fig. 6b). Supporting this model, PB size correlated positively with PIF5 stabilization and negatively with PIF5 degradation. A transition from small PBs in mCherry-PIF5/pif5-3 to large PBs in mCherry-PIF5/PBC switched the balance of PHYB signaling from degrading to stabilizing PIF5 (Figs. 2–4). Conversely, transferring mCherry-PIF5/PBC seedlings to dim light, which induced the transition from large PBs to small PBs, accelerated PIF5 degradation (Fig. 5). The current data suggest that localization of PIF5 to PBs impedes its functions in promoting hypocotyl elongation. This model corroborates the proposed function of PBs for sequestering PIF772,73. Enhancing the nucleoplasmic pool of mCherry-PIF5 in dim light promoted hypocotyl elongation (Fig. 5a, b). However, counterintuitively, the levels of both endogenous PIF5 and mCherry-PIF5 in mCherry-PIF5/PBC were reduced in the dim light compared to those in the strong light condition (Fig. 5f), indicating that the steady-state level of PIF5 does not correlate with its function in promoting hypocotyl elongation, likely due to enhanced degradation in the nucleoplasm. Another possibility is that PIF5 degradation is associated or coupled with its transcription activation activity. A similar mechanism was proposed for PIF3 as blocking PIF3 degradation in the hmr mutant attenuates the activation of its target genes60.
The function of PBs in stabilizing PIF5 may not be extrapolated to other PIFs, as PIFs are degraded via distinct mechanisms. The degradation of PIF1 is mediated by CRL4COP1/SPA and CRL1CTG10 with the F-box protein COLD TEMPERATURE GERMINATION 10 (CTG10) as the substrate recognition subunit69,74, whereas PIF3 is degraded by CRL3LRB and CRL1EBF. PBs were proposed to be required for PIF1 and PIF3 degradation7,24,29,58,59,61,62. However, it remains unclear whether PIF3 is degraded in PBs. Although PIF4 can also accumulate in the light36,46,55, PIF4 is ubiquitylated by the cullin3-based E3 ligase CRL3BOP1/2 using BLADE ON PETIOLE (BOP1/2) as the substrate recognition components75. When coexpressed with PHYB, FP-tagged PIF4, as well as PIF3 and PIF6/PIL2, localized with PHYB in biomolecular condensates in mammalian cells or protoplasts in a PHYB-dependent manner, supporting the idea that PHYB may be able to recruit PIF4 to PBs37,38. However, the PB localization of PIF4 in Arabidopsis has not been carefully studied, and the potential role of PBs in stabilizing PIF4 in the light remains to be experimentally verified.
A major difference separating this study from the previous investigations about PBs is the approach used to perturb PBs. Because disrupting PB assembly using loss-of-function mutants in PHYB and PHYB signaling could not uncouple PHYB signaling outputs in PBs and the surrounding nucleoplasm, here we perturbed the PB size by increasing PHYB abundance. If the LLPS of PHYB had driven PB formation, increasing the PHYB level would be expected to only enlarge the PB size without changing the concentrations of PHYB in the nucleoplasm4,63. Consistent with the PHYB LLPS model, increasing PHYB abundance enlarged the PB size (Fig. 1d, e). Most PHYB-CFP in PBs was immobile in gPBC-25 (Fig. 1g). This attribute of PBs was similar to the biomolecular condensates formed by PHYB alone in mammalian cells37. Consistently, increasing the PHYB level in PBC further reduced the fluorescence recovery kinetics and the mobile fraction of PHYB-CFP in PBs, indicative of a gel-like state (Fig. 1g). These results suggest that changes in PHYB abundance under the physiological PHYB concentration may alter both PB size and dynamics. Another unexpected result is that increasing PHYB abundance elicited a biphasic PIF5 response, implying that the nucleoplasmic PHYB increased in gPBC-25 to promote PIF5 degradation. One possibility is that at the physiological PHYB concentration in Col-0, PHYB condensation may form under the critical concentration via a complex mechanism beyond LLPS; in this scenario, like what was observed, overexpressing PHYB above the physiological PHYB concentration would alter both PB size and the nucleoplasmic PHYB concentration. Although we could not have revealed the function of PBs in stabilizing PIF5 without perturbing PBs using the PHYB overexpressing lines, it is important to note that a limitation of characterizing PB dynamics using PHYB overexpression lines would be the difficulty to completely exclude the possibility of a secondary effect on the signaling output due to PHYB overexpression.
This study reveals a PB-mediated light signaling mechanism in which PB formation spatially segregates two opposing PHYB signaling actions of PIF5 stabilization and degradation, which could enable a counterbalancing mechanism to titrate nucleoplasmic PIF5 and environmental responses (Fig. 6). We propose that this PB function represents a general function of biomolecular condensates to allow distinct variations of a cellular process or signaling pathway to coexist and interact to generate dynamically adjustable integrated outputs within a single subcellular space.
Methods
Plant materials, growth conditions, and hypocotyl measurements
The Arabidopsis Columbia ecotype (Col-0) was used throughout this study. The phyB-976, pifq77, and pif5-3 (SALK_087012)78 mutants, as well as the PBC line32, were previously described. The gPBC-25, gPBC-29, mCherry-PIF5/pif5-3 (#8 and #9) and mCherry-PIF5/PBC (#1 and #9) transgenic lines were generated in this study. Arabidopsis seeds were surface-sterilized and plated on half-strength Murashige and Skoog (½ MS) medium containing Gamborg’s vitamins (MSP06, Caisson Laboratories, Smithfield, UT), 0.5 mM MES pH 5.7, and 0.8% agar (w/v). Seeds were stratified in the dark at 4 oC for five days before treatment with specific light conditions and temperatures in an LED chamber (Percival Scientific, Perry, IA). Seedlings grown in the dark were exposed to 10 μmol m−2 s−1 FR light for 3 h after stratification to induce germination. Fluence rates of light were measured using an Apogee PS-200 spectroradiometer (Apogee Instruments, Logan, UT) and SpectraWiz® spectroscopy software (StellarNet, Tampa, FL). Images of representative seedlings were taken using a Leica M165 FC stereo microscope (Leica Microsystems, Deerfield, IL) and processed using Adobe Photoshop CC (Adobe, San Jose, CA). For hypocotyl measurements, seedlings were scanned using an Epson Perfection V700 photo scanner (Epson America, Los Alamitos, CA), and hypocotyl length was measured using NIH ImageJ software (http://rsb.info.nih.gov/nih-image/).
Plasmid construction and generation of transgenic lines
To generate the gPBC lines, PHYB genomic DNA containing a CFP fused in-frame before the stop codon was cloned into pJHA212G79. The resulting construct was transformed into phyB-9. To generate mCherry-PIF5/pif5-3 lines, a 2.3 kb genomic DNA fragment, encompassing the PIF5 promoter region, 5’ UTR, and the first two codons, was amplified using PCR and subcloned with 4× Myc, mCherry-2-L and the PIF5 coding sequence into pJHA212G79 containing the rbcS terminator using Gibson Assembly (New England Biolabs, Ipswich, MA). The resulting construct was transformed into pif5-3 using Agrobacterium-mediated transformation. To generate mCherry-PIF5/PBC lines, a 1.2 kb UBQ10 promoter was amplified using PCR and subcloned with 3× HA, mCherry-2-L and the PIF5 coding sequence into pJHA212B79 containing an rbcS terminator using Gibson Assembly. The resulting construct was transformed into PBC Agrobacterium-mediated transformation. The primers used for plasmid construction in this study are listed in Supplementary Table 1.
Protein extraction and immunoblotting
Total protein was extracted from Arabidopsis seedlings grown under the indicated conditions. For assessing the degradation kinetics of PIF5 and mCherry-PIF5, seedlings were vacuum infiltrated for 15 min in PBS containing 200 μM cycloheximide and then collected at the indicated time points. At least three independent degradation kinetics experiments were performed to calculate the average relative PIF5 or mCherry-PIF5 levels at each time point normalized to the PIF5 value at time zero. Plant tissues were ground in liquid nitrogen and resuspended in extraction buffer containing 100 mM Tris-HCl pH 7.5, 100 mM NaCl, 5% SDS, 5 mM EDTA pH 8.0, 20 mM dithiothreitol, 20% glycerol, 142 mM β-mercaptoethanol, 2 mM phenylmethylsulfonyl fluoride, 1× cOmplete™ EDTA-free Protease Inhibitor Cocktail (Sigma-Aldrich, St. Louis, MO), 80 μM MG132, 80 μM MG115, 1% phosphatase inhibitor cocktail 3 (Sigma-Aldrich, St. Louis, MO), 10 mM N-ethylmaleimide (NEM), 2 mM sodium orthovanadate (Na3OV4), 25 mM β-glycerophosphate disodium salt hydrate, 10 mM NaF, and 0.01% bromophenol blue. Protein extracts were boiled for 10 min and then centrifuged at 16,000 × g for 10 min at room temperature. Protein extracts were separated via 12% SDS-PAGE, transferred to a PVDF membrane, probed with the indicated primary antibodies, and then incubated with HRP-conjugated secondary antibodies. Rabbit anti-PIF5 polyclonal antibody (AS12 2112, Agrisera, Vännäs, Sweden) was used at a 1:1000 dilution. Mouse anti-PHYB monoclonal antibody (a gift from Dr. Akira Nagatani) was used at a 1:2000 dilution. Goat anti-mouse (1706516, Bio-Rad Laboratories, Hercules, CA) and goat anti-rabbit (1706515, Bio-Rad Laboratories, Hercules, CA) secondary antibodies were used at a 1:5000 dilution. Signals were detected via SuperSignal West Dura Extended Duration Chemiluminescent Substrate (Thermo Fisher Scientific, Waltham, MA). Western blots were quantified using ImageJ software (http://rsb.info.nih.gov/nih-image/). The relative level of the protein of interest was calculated by normalizing against the corresponding level of the actin control. The protein degradation kinetics curves were generated using data from at least three biological replicates.
Immunofluorescence staining
Whole-mount immunofluorescence staining was performed as described previously with the following modifications72,80. Seedlings were fixed in 4% (v/v) paraformaldehyde (15710, Electron Microscopy Sciences, Hatfield, PA), dehydrated, and mounted on slides72,80. All subsequent steps were performed in a 55 µL SecureSeal chamber (621505, Grace Bio-Labs, Bend, OR). Myc-tagged mCherry-PIF5 was detected using rabbit anti-Myc polyclonal antibodies (2272, Cell Signaling Technology, Danvers, MA) as the primary antibody at 1:100 dilution and donkey anti-rabbit AlexaFluor 555 antibodies (A31572, Thermo Fisher Scientific, Waltham, MA) as the secondary antibody at 1:1000 dilution. PHYB was detected using mouse monoclonal anti-PHYB antibodies (a gift from Akira Nagatani, 1:100 dilution) as the primary antibody and donkey anti-mouse AlexaFluor 488 antibodies (A21202, Thermo Fisher Scientific, Waltham, MA, 1:1000 dilution) as the secondary antibody. Nuclei were counterstained with 3.6 µM 4′,6-diamidino-2-phenylindole (DAPI). Samples were mounted using ProLong Gold Antifade Mountant (Thermo Fisher Scientific, Waltham, MA) and left to cure overnight in the dark before confocal analysis.
Seedling preparation for PB analysis using confocal microscopy
Seedlings were fixed following a previously described protocol with slight modifications81. Seedlings were fixed under vacuum with 1% (v/v) paraformaldehyde in PBS for 10 min. After quenching with 50 mM NH4Cl, the fixed seedlings were permeabilized with 0.2% Triton X-100 in PBS, and nuclei were stained with 3.6 µM DAPI in PBS for 10 min. Seedlings were washed with PBS before being mounted on a slide using Prolong Diamond Antifade Mountant (Thermo Fisher Scientific, Waltham, MA). The slides were left to cure overnight in the dark before being sealed with nail polish and stored at 4 oC. For live-cell imaging, seedlings were mounted using PBS. The slides were transported to the microscope in an aluminum foil-wrapped petri dish, and nuclei were imaged within 5 min after mounting.
Fluorescence microscopy and imaging analysis
For colocalization of mCherry-PIF5 and PHYB in mCherry-PIF5/pif5-3 using immunofluorescence staining, three-dimensional (3D) image stacks of individual nuclei of cotyledon pavement epidermal cells were imaged using a Zeiss LSM800 confocal microscope equipped with a ×100/1.4 Plan-Apochromat oil-immersion objective (Carl Zeiss AG, Jena, Germany). Alexa 488 was monitored using 488 nm excitation and 490–561 nm bandpass emission. Alexa 555 was monitored using 561 nm excitation and 560–630 nm bandpass emission. DAPI was monitored using 405 nm excitation and 410–470 nm bandpass emission. For colocalization mCherry-PIF5/PBC lines, 3D image stacks of individual nuclei of cotyledon pavement epidermal cells were imaged using a Zeiss LSM800 confocal microscope equipped with a ×100/1.4 Plan-Apochromat oil-immersion objective (Carl Zeiss AG, Jena, Germany). PHYB-CFP was monitored using 405 nm excitation and 410–470 nm bandpass emission. mCherry-PIF5 was monitored using 561 nm excitation and 560–630 nm bandpass emission. DAPI was monitored using 405 nm excitation and 410–470 nm bandpass emission. Deconvolution (nearest neighbor) was performed, and the maximum projection of image stacks (Z-stack interval 0.7 μm) was generated using Zeiss ZEN 2.3 software (Carl Zeiss AG, Jena, Germany). Representative images were exported as TIFF files and processed using Adobe Photoshop CC (Adobe, San Jose, CA). To measure the PB diameter of fixed mCherry-PIF5/PBC seedlings, the boundary of the PB peak was defined as the point where the fluorescence intensity was half of the peak value.
For PB analysis in gPBC-25, gPBC-29, PBC, and dark-grown mCherry-PIF5/PBC, 3D image stacks of individual nuclei from cotyledon epidermal or upper hypocotyl epidermal cells were imaged using a Zeiss Axio Observer Z1 inverted microscope equipped with a Plan-Apochromat 100x/1.4 oil-immersion objective and an Axiocam mono camera (Carl Zeiss, Jena, Germany). Fluorescence was detected using a broad spectrum X-Cite 120LED Boost high-power LED illumination system (Excelitas Technologies, Waltham, MA) and the following Zeiss filter sets: DAPI, excitation 365 nm, emission 445/50 nm/nm (Zeiss Filter Set 49); CFP, excitation 436/25 nm/nm, emission 480/40 nm/nm (Filter set 47 HE); mCherry, excitation 550/25 nm/nm, emission 605/70 nm/nm (Zeiss Filter set 43 HE), brightfield was used for the DIC image. Image stacks with a Z-step size of 0.24 µm were subjected to iterative classic maximum likelihood estimation deconvolution using Huygens Essential (Scientific Volume Imaging, Hilversum, Netherlands). The volume of PBs was quantified using the Huygens Object Analyzer (Scientific Volume Imaging, Hilversum, Netherlands). The PB partitioning of PHYB-CFP was measured using the Huygens Object Analyzer and 3D image stacks of PHYB-CFP and the DAPI-stained nucleus. Huygens Object Analyzer renders a 3D model of the PHYB-CFP signals in PBs and the entire nucleus that was defined by the DAPI signal. The PB-partitioning of PHYB-CFP was calculated as the percentage of PHYB-CFP in PBs versus the entire nucleus.
For FRAP experiments, seedlings were mounted in PBS and placed onto a Zeiss LSM800 confocal microscope with a C-Apochromat 63×/1.2 W autocorr M27 objective (Carl Zeiss AG, Jena, Germany). For the gPBC-25 and PBC experiments, PHYB-CFP was monitored using 405 nm excitation and 490–561 nm bandpass detector settings. Images were acquired at 2.7× magnification, the image format was 512 × 512 pixels, and the pinhole was set at 97 μm. One image was taken pre-bleach, followed by 6 iterations of photobleaching using the 405 nm laser at 85% power. Images after bleaching were collected using low laser intensities and were taken at 4 s intervals for 18 cycles. For the mCherry-PIF5/PBC experiments, PHYB-CFP was monitored using 405 nm excitation and 490–561 nm bandpass detector settings, and mCherry-PIF5 was excited with a 561 nm laser and observed using a 560–630 nm bandpass emission. Images were acquired at 2.7× zoom, the image format was 512 × 512 pixels, and the pinhole was 121 μm. One image was taken pre-bleach, followed by 8 iterations of photobleaching using the 405 nm laser at 50–60% power for PHYB-CFP and the 561 nm laser for mCherry-PIF5 at 85% power. Images after bleaching were collected using low laser intensities and were taken at 6 s intervals for 20 cycles. FRAP curve analyses were performed by setting the time of bleach to zero. Normalization was performed by taking the sum of the intensity of the ROI, subtracting the background, and setting the initial pre-bleach intensity to 1. Data were fitted to a one-phase exponential curve, and MF and t1/2 were calculated using Prism 10 (GraphPad Software, Boston, MA).
RNA extraction and quantitative real-time PCR
Total RNA from seedlings was isolated using a Quick-RNA MiniPrep kit with on-column DNase I treatment (Zymo Research, Irvine, CA). cDNA was synthesized using Oligo(dT) primers and 1 μg total RNA via a Superscript II First-Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA). Quantitative real-time PCR (qRT-PCR) was performed with iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) using a LightCycler 96 System (Roche, Basel, Switzerland). The transcript level of each gene was normalized to that of PP2A (At1G13320). The statistical analyses were performed using one-way analysis of variance (ANOVA) with post hoc Tukey’s HSD using Prism 10 (GraphPad Software, Boston, MA). The primers for qRT-PCR analysis used in this study are listed in Supplementary Table 2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Arabidopsis mutants and transgenic lines as well as plasmids generated during the current study are available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
Shaw, P. J. & Brown, J. W. S. Plant nuclear bodies. Curr. Opin. Plant Biol. 7, 614–620 (2004).
Mao, Y. S., Zhang, B. & Spector, D. L. Biogenesis and function of nuclear bodies. Trends Genet. 27, 295–306 (2011).
Belmont, A. S. Nuclear compartments: an incomplete primer to nuclear compartments, bodies, and genome organization relative to nuclear architecture. Cold Spring Harb. Perspect. Biol. 14, a041268 (2022).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Emenecker, R. J., Holehouse, A. S. & Strader, L. C. Biological phase separation and biomolecular condensates in plants. Annu. Rev. Plant Biol. 72, 17–46 (2021).
Liebl, M. C. & Hofmann, T. G. Regulating the p53 tumor suppressor network at PML biomolecular condensates. Cancers 14, 4549 (2022).
Van Buskirk, E. K., Decker, P. V. & Chen, M. Photobodies in light signaling. Plant Physiol. 158, 52–60 (2012).
Pardi, S. A. & Nusinow, D. A. Out of the dark and into the light: A new view of phytochrome photobodies. Front. Plant Sci. 12, 732947 (2021).
Chen, M. & Chory, J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 21, 664–671 (2011).
Hahm, J., Kim, K., Qiu, Y. & Chen, M. Increasing ambient temperature progressively disassembles Arabidopsis phytochrome B from individual photobodies with distinct thermostabilities. Nat. Commun. 11, 1660 (2020).
Legris, M. et al. Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354, 897–900 (2016).
Rockwell, N. C., Su, Y.-S. & Lagarias, J. C. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858 (2006).
Li, H., Burgie, E. S., Gannam, Z. T. K., Li, H. & Vierstra, R. D. Plant phytochrome B is an asymmetric dimer with unique signalling potential. Nature 604, 127–133 (2022).
Chen, M., Chory, J. & Fankhauser, C. Light signal transduction in higher plants. Annu. Rev. Genet. 38, 87–117 (2004).
Klose, C., Nagy, F. & Schäfer, E. Thermal reversion of plant phytochromes. Mol. Plant 13, 386–397 (2020).
Jung, J.-H. et al. Phytochromes function as thermosensors in Arabidopsis. Science 354, 886–889 (2016).
Delker, C., Quint, M. & Wigge, P. A. Recent advances in understanding thermomorphogenesis signaling. Curr. Opin. Plant Biol. 68, 102231 (2022).
Legris, M., Ince, Y, Ç. & Fankhauser, C. Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat. Commun. 10, 5219 (2019).
Wang, X. et al. A photoregulatory mechanism of the circadian clock in Arabidopsis. Nat. Plants 7, 1397–1408 (2021).
Más, P., Devlin, P. F., Panda, S. & Kay, S. A. Functional interaction of phytochrome B and cryptochrome 2. Nature 408, 207–211 (2000).
Kim, C. et al. Phytochrome B photobodies are comprised of phytochrome B and its primary and secondary interacting proteins. Nat. Commun. 14, 1708 (2023).
Kircher, S. et al. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11, 1445–1456 (1999).
Yamaguchi, R., Nakamura, M., Mochizuki, N., Kay, S. A. & Nagatani, A. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145, 437–445 (1999).
Van Buskirk, E. K., Reddy, A. K., Nagatani, A. & Chen, M. Photobody localization of phytochrome B is tightly correlated with prolonged and light-dependent inhibition of hypocotyl elongation in the dark. Plant Physiol. 165, 595–607 (2014).
Murcia, G., Enderle, B., Hiltbrunner, A. & Casal, J. J. Phytochrome B and PCH1 protein dynamics store night temperature information. Plant J. 105, 22–33 (2021).
Huang, H. et al. PCH1 integrates circadian and light-signaling pathways to control photoperiod-responsive growth in Arabidopsis. Elife 5, e13292 (2016).
Trupkin, S. A., Legris, M., Buchovsky, A. S., Tolava Rivero, M. B. & Casal, J. J. Phytochrome B nuclear bodies respond to the low red to far-red ratio and to the reduced irradiance of canopy shade in Arabidopsis. Plant Physiol. 165, 1698–1708 (2014).
Chen, M., Schwab, R. & Chory, J. Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc. Natl Acad. Sci. USA. 100, 14493–14498 (2003).
Chen, M. et al. Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141, 1230–1240 (2010).
Su, Y.-S. & Lagarias, J. C. Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell 19, 2124–2139 (2007).
Viczián, A. et al. Differential phosphorylation of the N-terminal extension regulates phytochrome B signaling. New Phytol. 225, 1635–1650 (2020).
Chen, M., Tao, Y., Lim, J., Shaw, A. & Chory, J. Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr. Biol. 15, 637–642 (2005).
Zhang, J., Stankey, R. J. & Vierstra, R. D. Structure-guided engineering of plant phytochrome B with altered photochemistry and light signaling. Plant Physiol. 161, 1445–1457 (2013).
Matsushita, T., Mochizuki, N. & Nagatani, A. Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424, 571–574 (2003).
Klose, C. et al. Systematic analysis of how phytochrome B dimerization determines its specificity. Nat. Plants 1, 15090 (2015).
Qiu, Y. et al. Mechanism of early light signaling by the carboxy-terminal output module of Arabidopsis phytochrome B. Nat. Commun. 8, 1905 (2017).
Chen, D. et al. Integration of light and temperature sensing by liquid-liquid phase separation of phytochrome B. Mol. Cell 82, 3015–3029.e6 (2022).
Beyer, H. M. et al. Red light-regulated reversible nuclear localization of proteins in mammalian cells and zebrafish. ACS Synth. Biol. 4, 951–958 (2015).
Huang, H. et al. PCH1 regulates light, temperature, and circadian signaling as a structural component of phytochrome B-photobodies in Arabidopsis. Proc. Natl Acad. Sci. USA. 116, 8603–8608 (2019).
Cheng, M.-C., Kathare, P. K., Paik, I. & Huq, E. Phytochrome signaling networks. Annu. Rev. Plant Biol. 72, 217–244 (2021).
Park, E. et al. Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 72, 537–546 (2012).
Park, E., Kim, Y. & Choi, G. Phytochrome B requires PIF degradation and sequestration to induce light responses across a wide range of light conditions. Plant Cell 30, 1277–1292 (2018).
Yoo, C. Y. et al. Direct photoresponsive inhibition of a p53-like transcription activation domain in PIF3 by Arabidopsis phytochrome B. Nat. Commun. 12, 5614 (2021).
Al-Sady, B., Ni, W., Kircher, S., Schäfer, E. & Quail, P. H. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23, 439–446 (2006).
Shen, Y., Khanna, R., Carle, C. M. & Quail, P. H. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145, 1043–1051 (2007).
Lorrain, S., Allen, T., Duek, P. D., Whitelam, G. C. & Fankhauser, C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323 (2008).
Shen, H., Moon, J. & Huq, E. PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 44, 1023–1035 (2005).
Pham, V. N., Kathare, P. K. & Huq, E. Dynamic regulation of PIF5 by COP1-SPA complex to optimize photomorphogenesis in Arabidopsis. Plant J. 96, 260–273 (2018).
Ni, W. et al. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 8, 15236 (2017).
Ni, W. et al. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344, 1160–1164 (2014).
Dong, J. et al. Light-dependent degradation of PIF3 by SCFEBF1/2 promotes a photomorphogenic response in Arabidopsis. Curr. Biol. 27, 2420–2430.e6 (2017).
Sheerin, D. J. et al. Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27, 189–201 (2015).
Lu, X.-D. et al. Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant 8, 467–478 (2015).
Saijo, Y. et al. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17, 2642–2647 (2003).
Qiu, Y., Li, M., Kim, R. J.-A., Moore, C. M. & Chen, M. Daytime temperature is sensed by phytochrome B in Arabidopsis through a transcriptional activator HEMERA. Nat. Commun. 10, 140 (2019).
Qiu, Y. et al. RCB initiates Arabidopsis thermomorphogenesis by stabilizing the thermoregulator PIF4 in the daytime. Nat. Commun. 12, 2042 (2021).
Kaiserli, E. et al. Integration of light and photoperiodic signaling in transcriptional nuclear foci. Dev. Cell 35, 311–321 (2015).
Bauer, D. et al. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16, 1433–1445 (2004).
Galvão, R. M. et al. Photoactivated phytochromes interact with HEMERA and promote its accumulation to establish photomorphogenesis in Arabidopsis. Genes Dev. 26, 1851–1863 (2012).
Qiu, Y. et al. HEMERA couples the proteolysis and transcriptional activity of PHYTOCHROME INTERACTING FACTORs in Arabidopsis photomorphogenesis. Plant Cell 27, 1409–1427 (2015).
Yoo, C. Y. et al. Phytochrome activates the plastid-encoded RNA polymerase for chloroplast biogenesis via nucleus-to-plastid signaling. Nat. Commun. 10, 2629 (2019).
Yang, E. J. et al. NCP activates chloroplast transcription by controlling phytochrome-dependent dual nuclear and plastidial switches. Nat. Commun. 10, 2630 (2019).
Klosin, A. et al. Phase separation provides a mechanism to reduce noise in cells. Science 367, 464–468 (2020).
Sharma, A. et al. UVR8 disrupts stabilisation of PIF5 by COP1 to inhibit plant stem elongation in sunlight. Nat. Commun. 10, 4417 (2019).
Balleza, E., Kim, J. M. & Cluzel, P. Systematic characterization of maturation time of fluorescent proteins in living cells. Nat. Methods 15, 47–51 (2018).
Niwa, Y., Yamashino, T. & Mizuno, T. The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol. 50, 838–854 (2009).
Banani, S. F. et al. Compositional control of phase-separated cellular bodies. Cell 166, 651–663 (2016).
Jang, I.-C., Henriques, R., Seo, H. S., Nagatani, A. & Chua, N.-H. Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22, 2370–2383 (2010).
Zhu, L. et al. CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat. Commun. 6, 7245 (2015).
Paik, I. et al. A phyB-PIF1-SPA1 kinase regulatory complex promotes photomorphogenesis in Arabidopsis. Nat. Commun. 10, 4216 (2019).
Chen, H. et al. Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18, 1991–2004 (2006).
Willige, B. C. et al. PHYTOCHROME INTERACTING FACTORs trigger environmentally responsive chromatin dynamics in plants. Nat. Genet. 53, 955–961 (2021).
Xie, Y. et al. Phytochrome B inhibits the activity of phytochrome-interacting factor 7 involving phase separation. Cell Rep. 42, 113562 (2023).
Majee, M. et al. KELCH F-BOX protein positively influences Arabidopsis seed germination by targeting PHYTOCHROME-INTERACTING FACTOR1. Proc. Natl Acad. Sci. USA. 115, E4120–E4129 (2018).
Zhang, B. et al. BLADE-ON-PETIOLE proteins act in an E3 ubiquitin ligase complex to regulate PHYTOCHROME INTERACTING FACTOR 4 abundance. Elife 6, e26759 (2017).
Reed, J. W., Nagpal, P., Poole, D. S., Furuya, M. & Chory, J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5, 147–157 (1993).
Leivar, P. et al. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18, 1815–1823 (2008).
Khanna, R. et al. The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19, 3915–3929 (2007).
Yoo, S. Y. et al. The 35S promoter used in a selectable marker gene of a plant transformation vector affects the expression of the transgene. Planta 221, 523–530 (2005).
Feng, C.-M., Qiu, Y., Van Buskirk, E. K., Yang, E. J. & Chen, M. Light-regulated gene repositioning in Arabidopsis. Nat. Commun. 5, 3027 (2014).
Yoo, C. Y., Williams, D. & Chen, M. Quantitative analysis of photobodies. Methods Mol. Biol. 2026, 135–141 (2019).
Acknowledgements
We thank Akira Nagatani (Kyoto University) for providing anti-PHYB antibodies. We thank Elise Pasoreck for valuable suggestions and comments on the manuscript. We are grateful to Xuemei Chen (Peking University) for her support of D.F. in a collaborative project on thermomorphogenesis. This work was supported by National Institute of General Medical Sciences grant R01GM087388 to M.C.
Author information
Authors and Affiliations
Contributions
R.J.K., D.F., J.H., K.K., J.D. and M.C. conceived the original research plan; M.C. supervised the experiments; R.J.K., D.F., J.H., K.K., J.D. and M.C. performed the experiments; R.J.K., D.F., J.H., K.K., J.D. and M.C. analyzed the data; R.J.K., D.F., J.H. and M.C. wrote the article with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Monika Chodasiewicz and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, R.J.A., Fan, D., He, J. et al. Photobody formation spatially segregates two opposing phytochrome B signaling actions of PIF5 degradation and stabilization. Nat Commun 15, 3519 (2024). https://doi.org/10.1038/s41467-024-47790-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-47790-8
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.